INTRODUCTION
Linamarase (β-glucosidase) is a hydrolytic enzyme. It degrades the glycolytic bond betweeen β- glucose molecule and the chiral carbon atom linked to the nitrile group of linamarin present in cassava. Linamarase (β-glucosidases) is economically very important because of its role as a detoxification agent for cyanogenic glucosides thereby improving food safety. Even though it can be produced from cassava (Manihot esculenta Crantz) tubers, the yields of native linamarase from this source is usually too low (Nok and Ikediobi, 1999). Linamarase from cassava is expensive and it is economically unwise to produce the enzyme from edible tubers. The problems associated with native linamarase (β-glucosidases) (CNLIN) used for detoxification include low enzyme concentration, limited spectrum of substrates activity and sensitivity to environmental factors such as pH and temperatures. Over the years however, humans have realized that selective propagation of animals and plants of desirable traits can improve and increase enzyme production in yield and quality. β-glucosidases of microbial origin may represent a possible solution to the problem. This approach has also been extended to bacteria and fungi for the increased downstream products development for the pharmaceutical, food and brewing industries. Yeast (Saccharomyces cerevisiae) for example, has long been used for the production of high yielding metabolites and other food enzymes including amylases and zymase for hydolysis of starch and fermentation of glucose into ethanol respectively (Mach and Zeilinger, 2009). Some quantities of linamarase were for the first time extracted from yeast by Ikediobi and Ogunda (1985) and Okafor and Ejiofor (1985). Yeast like other microorganisms has been extensively used in genetic engineering for the production of single- cell proteins, enzymes, hormones and vitamins. This is because of their low generation times and ease of manipulation. The kinetic data of genetically engineered β-glucosidase on linamarin extracted from cassava will provide insight into the mechanism of action of the enzyme while providing the parameters necessary for predictive purposes. Such predictions which are lacking can be useful tools for fermentation process optimization for the degradation of cyanogenic glucosides in cassava-based food systems. The research objective of this study was to characterise genetically engineered β-glucosidase from Saccharomyces cerevisiae by its action on cassava linamarin using commercial native linamarase as the control for this study. The approach is to boost products transformation and development of cassava-based food products that can contribute to safety global nutrition and food security, increase international export market for national income earning from cassava-based food products.
MATERIALS AND METHODS
Materials:
Enzyme samples: Commercial Native Linamarase (CNLIN) used as the control for this study was purchased from Sigma Co. Lousina, USA. Genetically Engineered β-glucosidase (GELIN) was obtained from the Department of Food Science and Technology University of Agriculture Makurdi. Crude extract from Saccharomyces cerevisiae (GELIN0) was further purified using Carboxy methyl cellulose (GELIN1), Diethyl-amino-ethyl- sephadex (GELIN2) and Diethyl-amino-ethyl-cellulose (GELIN3)
Production of Linamarin(Substrate): Linamarin (100g) was extracted from about 2kg tubers of the bitter wild cassava (Manihot esculenta pohr) variety TSM-TRF-2005035 obtained from Tse-Akaa Village Mbalagh, Makurdi, Benue State, Nigeria. It was processed using the method described by Ikediobi and Ogunda (1985).
Linamarin: One year old cassava tubers were harvested washed with tap water and promptly frozen overnight at -10oC. About 800g of frozen cassava parenchyma tissues were sliced with stainless steel knives and homogenized with 160ml of chilled 0.1M phosphoric acid solution. The resultant slurries were filtered rapidly using glass wool and the filtrate centrifuged (1000 rpm) for 5min.The resultant filtrate was centrifuged at 5000rpm for 5min and the supernatant adjusted to pH 8.0 followed by re-centrifugation at 5000rpm for 8 mins. After decanting, the solid residue was air-dried to obtain about 0.82g of white substrate (mp.143oC).This was stored at 4 oC and subsequently used for characterization of activity kinetic profiles.
Buffer Solutions and Analytical grade Reagents: Buffer solutions and reagents were prepared for the study using standard methods.
Kinetic studies: The ability of Commercial Native Linamarase (CNLIN) purchased as control and the Genetically Engineered Linamarase (GELIN) extracts from Saccharomycescerevisiae to hydrolyse cassava linamarin to hydrocyanic acid (HCN) released within a fixed time was challenged. The spectrophotometric method used was used for the estimation of hydrocyanic acid (HCN) released within a fixed time as described by Onyike et al., (2001).
Characterization of physical and activity kinetic profiles
Characterization of commercial native linamarase (CNLIN) and genetically engineered linamarase (GELIN) from yeast (Saccharomyces Cerevisiae) was carried out in this study with the following materials and method:
Materials: Vertical polyacylamide gradient slabs (13-24 %), 2-methylmercapto ethanol, Coomassie G brilliant blue, Mwt. calibration standards (phosphorylase, b(94,000), bovine serum albumin(67,000), ovalbumin (43,000), carbonic anhydrase(30,000), soybean trypsin inhibitor(20,000), lactalbumin (14,000) and fragments of myoglobin(17, 200; 14,6oo; 8,240; 6,380 and 2,560). Fractogel TSK HW 50(F) column (90×2.5cm), 0.1 %(v/v)TFA,70% (V/V) Ethanol, appropriate sodium acetate buffer, stop clock, spectrophotometer, chromotographic Column, Kontron Liquimat111 amino acids Analyser 6M HCl, weighing balance, thermometer test tubes, performic acid.
Procedure for characterization of physical profiles
Vertical Soduim dodecyl sulphate polyacylamide gel electrophoresis Method for molecular weight (Mr.) determination and separation of enzyme polypepetides was done under non-reducing and reducing condition using 2-methylmercapto ethanol. Vertical SDS-PAGE was performed on polyacylamide gradient slabs (13-24%. Gels were stained with Coomassie G brilliant blue. Mr. calibration standards were: phosphorylaseb(94,000), bovine serum albumin(67,000), ovalbumin(43,000), carbonic anhydrase(30,000), soybean trypsin inhibitor (20,000), lactalbumin (14,000) and fragments of myoglobin (17,200; 14,600; 8,240; 6,380 and 2,560).
Molecular-Exclusion/Gel Filtration Chromatographic Separation method
This was performed on a Fractogel TSK HW 50(F) column (90×2.5cm equilibrated 0.1 %(v/v) TFA,70% (V/V) Ethanol. Enzyme fractions were eluted with appropriate buffer at 25 ml/h. Fractions were detected at 224-280nm, collected and subjected to Mr. using standard Mr. markers for calibration of the column.
Kontron Liquimat 111 aminoacids Analyser
Analytical determination and separation of amino acids was applied in evaluting s-containing amino acid. Pure samples each (0.5mg) were hydrolyzed under vacuum in 6M HCl for 24hr. at 110oC in sealed tubes. Sulphur amino acids were analysed after performic acid oxidation to methionine sulfone and cysteic acid. Amino acids compositions were determined using a Kontron Liquimat111 Analyser.
Estimation of Insoluble proteins: The insoluble proteins were estimated as described by Nok & Ikediobi(1999) An automated refrigerated centrifuge with Lowrys based principle was applied in the estimation of insoluble protein impurity.
Procedure for characterization of activity kinetic profiles: The activity profile kinetics of commercial linamarase (CNLIN) genetically engineered linamarase (GELIN) were derived by actions of enzymes at varying pH 1-14 on experimental linamarin as substrates. The Spectrophotometric method for estimation of hydrocyanic acid (HCN) released within a specified time of ten minutes as described by Onyike et al., (2001) was used for the investigation for the evaluation of the activity kinetic profiles parameters. In this procedure Enzyme aliquots (0.1ml) were prepared and added to tubes containing 0.5ml of 5µmol linamarin. The tubes were incubated for 30min at 3O0C at varying pH 1-14 and the reaction was stopped by adding 1ml of 0.2NaOH and 1ml of 0.2MHCl which was added to neutralize. Chloramine T(1ml of 1%) was added and after 1min., 3ml barbituric acid/pyridine reagent was added. The volume adjusted to 25ml distilled water followed by measurement of absorbance of the pink color at 420 nm.HCN was used to calibrate the absorbance values. The volume was increased to 25ml and the absorbance of the pink color was measured. H2SO4 was used to calibrate the absorbance values. The hydrocyanic acid (HCN) present in each tube was determined by spectrophotometric method. The absorbance measured in the enzyme assaywas used to calculate the total activity in units per ml of enzyme solution, where one unit is defined as that which produced 1micro mole of HCN per minute at 3O0C. The total and specific activities, purity fold, yield and purification efficiency of genetically engineered linamarase were as described by Onyike et al., (2001). Activity kinetic profiles parameters applied for the characterization of the enzyme samples include; total activity, specific activity, purity fold, yield and purification efficiency. Calculation of data from total activities and impurity of enzyme samples are shown mathematically in equations (1- 5) as described by Nok and Ikediobi (1999) and Onyike et al., 2001.

Statistical Analysis The tests for significant (p≤0.05) difference in the purification profile, enzyme and activity kinetics parameters at ambient temperature, and at varying pH, temperature and purity fold were calculated with the multiple comparison range method of Krammer and Twigg (1970) and Gupta (1979).
RESULTS AND DISCUSSION
Data representing the structural characteristics of commercial native linamarase (CNLIN) and genetically engineered linamarase (GELIN) from yeast (Saccharomyces cerevisiae) are shown in Table I. The ionic charge of (0) was identical in CNLIN and GELIN0, whereas GELIN1 was positively charged. Both GELIN2 and GELIN3 were negatively charged. All the enzymes CNLIN, GELIN0, GELIN1, GELIN2 and GELIN3 contained methionine corresponding respectively to 2, 15, 23, 25 and 26%. The enzymes similarly contained cysteine in the proportions of 3, 13, 14, 17 and 20% respectively. Molecular weights estimated in Dalton units were respectively 160,100, 260,000, 240,000, 230,000 and 220,000 using Sodium dedocyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) techniques, whereas the mol.wt estimated by gel filtration chromatography were 160, 600, 260, 100, 240, 100, 230, 100 and 220, 200. The impurity (mg/100g sample) in enzymes evaluated 2.42, 3.52, 0.9, 0.6 and 0.4 respectively. Each fraction had three isozymes.The impurities(mg/100g) in the fractions were 2.42(CNLIN), 3.52(GELIN0), 0.9 (GELIN1), 0.6(GELIN2) and 0.4(GELIN3) showing increased purity of purified enzymes. The value for GELIN0 was not (p< 0.05) higher than 2.42mg/100g of commercial native linamarese (CNLIN). The presence of two active isoenzymes in commercial native linamarse (CNLIN) was earlier reported by Nok and Ikediobi (1999). Later reports by Wither et al.(2002) revealled that a good number of isozymes are responsible for the wide spectrum of substrates specificity The molecular weight(Dalton units) of 22,000- 26,000 , for GELIN3-GELIN0 were not highly significant (p<0.05) different form the molecular weight of carbonic anlydrase of 30,000 daltons but however, significantly different (p> 0.05) from the 63,000 of phosphorylase (b) 94,000 serum albumin 67,000, ovalbumin 43,000 The values for the GELIN were however, higher than trypsin inhibitor (20,000) lacalbumin 14,000 and fragements of myoglobin 17,200, 14 600 , 8,200, 6,380 and 2,5600) making the GELIN0– GELIN3 and CNLIN to be classified as medium molecular weight enzyme fractions. Molecular weight determination by Cicek et al., (1998) showed that linamarase was in the range of 57 to 63 KD. The methionine and cystiene contents ranged from15-26 % and 13-20% respectively. (GELIN0- GELIN3) of sulphur amino acid evaluated were significantly higher (p< 0.05) than the commercially native linamarase (CNLIN) indicating the contribution of genetic engineering in incorporating the surphur bridges in enzymes to thermo-stable and probably pH tolerant structural characteristics produced of the cloned enzymes (Archer, 2006).
 |
TABLE I: Physical characteristics of commercial native and genetically engineered Linamarase from yeast (Saccharomyces cerevisiae) Click here to View table |
Characterization of activity kinetic profiles
Characterization of activity kinetic profiles related to the degradation of linamarin is discussed in section. The degradation of linamarin by the commercial and genetically engineered enzymes investigated were related to characterization activity kinetic profiles related to parameters such as; optimum pH , temperature, total activity, specific activity, purity fold and yield. The performance of GELIN3 were significantly (P< 0.05) higher than CNLIN. Table II represents the computed values in equations 1-5 validated the activity profile kinetic models in describing the degradation of linamarin by both native and genetically engineered linamarase from Saccharomyces cerevisiae. The results are in accordance to those determined by Onyike et al., (2001), Nok and Ikediobi (1999) showing that the total activity was in the range of 9-14micromole HCN released per minute.
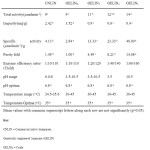 |
Table II: Activity kinetic profiles of native and genetically engineered linamarase from yeast (Saccharomycescerevisiae) in relation to action on linamarin Click here to View table |
In each of the cases, the examined enzymes had a common optimum pH and temperature in agreement with the previous studies of Karl – Joseph et al., (2002). The third degree purified genetic enzyme perfumed better than all enzyme factions. The commercial native linamarase performed within a narrow pH range of 6.8 – 6.9 in the degradation of linamarin. The native enzyme had no action on linamarin at pH 3.5 and 10.5. The genetic enzymes had a broad specificity activity hydrolyzing the experimental linamarin within the range of pH 3.5 and 10.5. This was in agreement with the work of Pertruccivlo et al., (1999) where genetic enzymes had a broad spectrum of action capable of hydrolyzing linamarin at a wide range of tolerable pH 1-14. The specific activity in micromole minute per gram range from 2.56 to 35 showing that purification in this study was very perfect. This was in agreement with the studies of Ekisttikul et al., (2007). The purity fold was within the range of 1 – 13.7 agreeing with the work of Ekisttikul et al., (2007). The enzyme Efficiency was ranged from 10 to 15.5 indicating the efficiency purification method applied in the removal of contaminating proteins for the genetically engineered enzymes. This was in agreement with the work Cicek et al., (1998). The similarity of results obtained by degradation of linamarin by different enzyme fractions could be as a result of sequence similarity of all the genetically engineered samples. On the whole, all the genetically engineered enzyme samples shared almost 100% sequence similarity while the commercial native enzyme shared only 30% sequence similarity with the genetic enzymes (the GELIN groups). This observation is in agreement with the Ciek et al., (1998). Which showed that genetically engineered b – glucosidases shared a common sequence identity of at least 70%
Conclusion
Recombinant DNA technology has been used to express and produce linamarase (β-glucosidase) comprising of three isoenzymes from Saccharomycecerevisiae. Ion-exchange chromatography techniques were capable of purifying the enzyme for activity enhancement. The genetically engineered linamarase from Saccharomyces cerevisiae exhibited high specificity activity by acting on the cyanogenicglucoside (linamarin) studied. Total activity increased with increased degree of purification at pH 6.8 and 35oC. The degradation of the linamarin by the genetically engineered linamarase obeyed characteristics of protein enzymes. The genetically engineered linamarase from Saccharomyces cerevisiae is recommended for detoxification of cyanide in cassava-based food products.
REFERENCES
- Acher D. B. and Peberdy J. F. The molecular biology of secreted enzyme production by fungi. Crit. Rev Biotechnol., 17:273–306 (2006).
CrossRef - Ikediobi C.O. and Ogundu E.C. Screening of some fungal isolate for linamarase production. NIFOJ Vol 3(1,2 and 3):165-167(1985)
- Ikediobi C.O. and Onyike E. The use of linamarase in garri production. Process Biochem. 17: 2-5(2002)
- Kramer A and Twigg B. A.(1970) Quality Control for the Food Industry. 3rd.Edition. Westport, Connecticut. The Avi Publishing Company Inc.pp.155-205 (1970)
- Nok J.N. and Ikediobi C.O. Some properties of linamarase from cassava (Manihot esculenta Crantz) cortex. J. Food Biochem.14: 477-489 (1999)
CrossRef - Petrucciioli M., Brimer R., Cicalini A.R., Pulci V. and Federici F. Production and properties of the linamarase and amygdalase activities of Penicillium auranthiogrisum. Biosci. Biotechnol. Biochem. 635 805-812 724 (1999)
- Cicek M. and Essen H. Structural abd expression of dhurrianse (β glucosidase from sorghum) Plant Physiol. 116: 1469-1478 (1998)
CrossRef - Ejiofor M.A.N. and Okafor N. Studies on microbial breakdown of linamarin in fermenting cassava NIFOJ. Vol.3(1,2and 3):153-158(2005)
- Eksittikul T. and Chulavalnatol M. Characterization of cyanogenic β-glucosidase from cassava(Manihot esculenta Crantz). Archiv Biochem Biophys 266:263-269 (2007)
CrossRef - Elamayerhi H. Mechanisms of pellet formation of Aspergillus niger with additives. J Ferment Technol;53:722–9 (2007)
- Gupta C.B. An Introduction to Statistical Methods.8th. Edition.pp.424-480. New Delhi. G. Vikas Publishing House, PVT Limited (1979)
- Ikediobi C.O. and Ogundu E.C. Screening of some fungal isolate for linamarase production. NIFOJ Vol 3.Nos 1,2 and 3 p.165-167(1985)
- Karl-Joseph D., Sauter A., Wichart R., Messdaghi D. and Harttury W. Extracellular β-glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. Journal of experimental food chemistry. Vol. 51( 346):937-944 (2000)
- Mach R. L. and Zeilinger S. Regulation of gene expression in industrial fungi: Trichoderma. Appl Microbiol Biotechnol ;60:515–22 (2009)
CrossRef - Nok J.N. and Ikediobi C.O. Some properties of linamarase from cassava (Manihot esculenta Crantz) cortex. J. Food Biochem.14: 477-489 (1999)
CrossRef - Onyike, E. Ukoha, A. and Ikediobi C.O. Isolation and Characterization of Linamarase from Dried Cassava (Manihot esculenta Crantz) Cortex. Nig. J. of Biochem and Mol. Biol. Vol.16(3): 739-785 (2001)
- Petrucciioli M., Brimer R., Cicalini A.R., Pulci, V. and Federici F. Production and properties of the linamarase and amygdalase activities of Penicillium auranthiogrisum P.35.Biosci.Biotechnol.Biochem. 635 805-812 (1999)
CrossRef - Withers, S.G. Identification of active- site residues in glucosidase. In Biological and Biotechnological.Application of ES1-MS, ACS Symposuim Series vol.619 (snyder, A.,P ),pp.365-380 (2002)

This work is licensed under a Creative Commons Attribution 4.0 International License.





