Introduction
The capacity of Staphylococcus aureus to acquire antibiotic resistance is among the most crucial characteristics to take into account. Staphylococcus aureus is a serious public health problem because it has developed resistance to several medications over time 1. The abuse and overuse of antibiotics frequently contribute to the development of resistance. Vancomycin resistance is among Staphylococcus aureus’ most prominent resistance mechanisms. Historically, illnesses brought on by bacteria resistant to other antibiotics have been treated with vancomycin as a last option. It works very well against Staphylococcus aureus and other Gram-positive bacteria. The medical community is concerned about the rise in vancomycin-intermediate Staphylococcus aureus (VISA) and vancomycin-resistant Staphylococcus aureus (VRSA) strains.The primary mechanism of vancomycin resistance in Staphylococcus aureus is the acquisition of distinct genes referred to as the vanA, vanB, vanC, vanD, and vanE genes. 2. These genes produce enzymes that alter the bacterial cell wall’s structure, preventing vancomycin from attaching to the cell wall, which is the drug’s intended target. The antibiotic thereby loses its ability to combat the resistant strains. The development of VISA and VRSA strains complicates the treatment of infections caused by Staphylococcus aureus. Alternative antibiotics, which can be less effective or have more adverse effects, are the only option available to healthcare practitioners. To treat these infections, combination medications or surgical procedures may be required in some circumstances. The creation of novel medicines, enhanced infection control strategies, and the encouragement of appropriate antibiotic usage are all part of the fight against vancomycin-resistant Staphylococcus aureus. Healthcare facilities have also implemented stringent hygiene protocols to prevent the spread of these resistant strains within healthcare settings. Additionally, research is ongoing to better understand the mechanisms of resistance and to find novel treatment options 3,4. Vancomycin is now used to treat people allergic to semisynthetic penicillin or cephalosporins, as well as infections brought on by these pathogens5. Vancomycin resistance in bacteria is mediated by van gene groups that are found in pathogens (such as S. aureus), glycopeptide-producing actinomycetes. Vancomycin resistance is classified into several gene clusters based on the DNA sequence of the ligase van gene homologues that encode the key enzyme for the synthesis of D-alanyl–D-lactate (D-Ala–D-Lac) or D-alanyl–D-serine (D-Ala–D-Ser). At least 11 van gene clusters that confer vancomycin-resistance, responding for VanA, VanB, VanC. High-level vancomycin resistance is frequently caused by the genes vanA, vanB, vanD, van F, vanI, and vanM, which encode D-Ala:D-Lac ligases. Transposon Tn1546 carries the original vanA gene cluster. History of previous vancomycin usage the majority of patients reported using vancomycin at least three months before being isolated from VRSA. It appears that the vanA gene cluster-induced drug resistance is selectively influenced by vancomycin illnesses that precede VRSA infection Patients with VRSA infections typically had many comorbid conditions, such as diabetes, end-stage kidney failure, and surgical or gangrenous wounds. Vancomycin-resistant plasmids were transferred from VRE to S. aureus more easily by biofilms that developed in wounds or catheters. A potent antibiotic against VRSA isolates may be treated with a variety of medicines. Vancomycin-resistant plasmids are genetic elements that can confer resistance to the antibiotic vancomycin. Vancomycin is a potent antibiotic commonly used to treat serious infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA) and Enterococcus sp. The development and spread of vancomycin resistance are concerning because it limits the effectiveness of this critical antibiotic. Plasmids are small, circular pieces of DNA that exist independently of the chromosomal DNA in bacteria. They can carry genes that provide various advantages to the bacteria, including antibiotic resistance genes. In the case of vancomycin resistance, certain plasmids carry genes that encode vancomycin resistance mechanisms. These resistance genes can be transferred between bacteria, facilitating the spread of resistance. There are different mechanisms by which plasmids can confer vancomycin resistance:1. VanA, VanB, and VanC Genes: These genes encode enzymes that modify the peptidoglycan cell wall of bacteria, preventing vancomycin from binding and disrupting the cell wall. The study aimed to examine the major genes (VanA,VanB,Vanc) for vancomycin resistance in staph from various clinical specimens.
Methodology
Out of 160 total specimens obtained from burns, blood, and urine, the study focused on 40 staph aureus specimens with relative percentages of 30.7%, 25%, and 18.18%. Culturing and biochemical testing were used to identify isolates. In order to isolate Staph aureus, from various specimens at numerous hospitals in Hilla City were collected. The clinical microbiologists and accompanying physicians determined what was deemed clinically relevant.
Fostering using sterile loops dispersed over the surface of the agar medium and a 24-hour incubation period at 37°C, all specimens were grown on various media, including blood agar, MacConkey agar, and Mannitol salt agar, for the purpose of identifying Staph aureus as shown in Figure (2), microscopic analysis following the bacterial growth on blood, MacConkey, and mannitol salt agar, the colonies’ size, shape, texture, and arrangement were noted. A single colony was taken out, stained with Gram stain, and evaluated using oil emersion, biochemical assays, under a light microscope (100x). Examine catalase: Favorable Positive coagulase test: Separate S. aureus from coagulase-negative Phyloglococcus. CONS are further differentiated on the basis of novobiocin sensitivity test (S. epidermidis is sensitive, whereas S. saprophyticus is resistant.(Staphylococcus aureus)Capsule Non-Capsulated Catalase Positive (+ve)Citrate Positive (+ve)Coagulase Positive (+ve)Gas Negative (-ve)Gelatin Hydrolysis Positive (+ve)Gram Staining Positive (+ve)H2S Negative (-ve)Hemolysis Positive (+ve)- BetaIndole Negative (-ve)Motility Negative (-ve)MR (Methyl Red) Positive (+ve)Nitrate Reduction Positive (+ve)OF (Oxidative-Fermentative)Fermentative Oxidase Negative (-ve)Pigment Mostly Positive (+ve)PYR Negative (-ve)Shape Cocci Spore Non-Sporing Urease Positive (+ve)VP (Voges Proskauer)Positive (+ve)Fermentation of Arabinose Negative (-ve)Cellobiose Negative (-ve)DNase Positive (+ve)Fructose Positive (+ve)Galactose Positive (+ve)Glucose Positive (+ve)Lactose Positive (+ve)Maltose Positive (+ve)Mannitol Positive (+ve)Mannose Positive (+ve)Raffinose Negative (-ve)Ribose Positive (+ve)Salicin Negative (-ve)Sucrose Positive (+ve)Trehalose Positive (+ve)Xylose Negative (-ve)Enzymatic Reactions Acetoin Production Positive (+ve)Alkaline Phosphatase Positive (+ve)Arginine Dehydrolase Positive (+ve)Hyalurodinase Positive (+ve)Lipase Positive (+ve)Ornithine Decarboxylase Negative (-ve).
The AST for 40 isolates using (8 )antibiotics (cefepime (30 μg), ceftriaxone (30 μg), clindamycin (2 μg), erythromycin (15 μg), chloromaphenicol (30 μg), imipenem (10 μg), tetracycline (30 μg) and nitrofurane(10 μg) were distinguished founded on CLSI,2022. The genes utilized in this investigation are mentioned in Table 1 below.
Table 1: Genes used in this study
|
Genes Name |
Sequence of genes(5’_3′) |
Amplcone size (bp) |
References |
|
VanC |
5′-GAAAGACAACAGGAAGACCGC-3 |
510bp |
20 |
|
5′-TCGCATCACAAGCACCAATC-3′ |
|||
|
VanA |
5′-AATGTGCGAAAAACCTTGCG-3′ |
520 bp |
19 |
|
5′-CCGTTTCCTGTATCCGTCC-3′ |
|||
|
VanB |
5′-GCTCCGCAGCCTGCATGGA-3′ |
580 bp |
20 |
Results and Discussions
Staph aureus Detection and Isolation
40 isolates (or 25%) out of the 160 samples (40 blood, 65 burns, and 55 urine samples) were found to be staph aureus.. However, 40 out of the 160 isolates staph aureus in 25% ,(37%) as different bacterial species. While negative culture (38%).
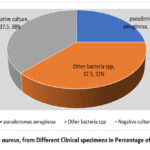 |
Figure 1: Staph aureus, from Different Clinical specimens in Percentage of Isolation rate . |
 |
Figures 2: Showed staph aureus on mannitol salt agar |
Distribution of Bacterial Isolates Among Different Specimens
Staph aureus isolates had been glued with numerous percentages from medical specimens, as shown in table (2). Distribution of staph aureusisolates among clinical specimens. The distribution of bacterial isolates among different specimens can vary widely depending on the type of specimens being analyzed and the specific study or context. Bacterial isolates refer to the types of bacteria that have been identified or isolated from various sources, such as clinical samples, environmental samples, or research studies. The distribution of bacterial isolates is an important aspect of microbiology and can provide insights into the types of bacteria present in a particular environment or the causes of infectious diseases. Blood: Bacterial isolates from blood specimens are typically associated with bloodstream infections. Common isolates include Staphylococcus aureus, Escherichia coli, Urine: Urinary tract infections (UTIs) often involve bacteria like Escherichia coli, Klebsiella sp., and Enterococcus sp. Burn swabs: Bacterial isolates from wound infections can include Staphylococcus aureus, Pseudomonas aeruginosa, and various Streptococcus sp.
Table 2: Distribution of bacterial isolates among different specimens.
|
Type of specimens |
N°of specimens |
N°of Isolates |
Percentage |
|
Burn swab |
65 |
20 |
40.625% |
|
Blood |
40 |
10 |
25 % |
|
Urine |
55 |
10 |
34.375 % |
|
Total |
160 |
40 |
100% |
Rendering to the study’s findings, out of 160 samples, 40 (25%) isolates were found to be staph aureus and they were distributed among burn swabs 20 (40.625%), wound swabs 10 (25%), urine 10(34.375%). According to Hosu investigations 9, they were find wound swabs 43.3% and burns 3.7% while another study by 10 showed 21.6%positive culture for staph aureus including (10% urine, 22% blood, 40% burn) and study by 11 fineding 32(26.6%) out of 120 specimens ( 28burns, 4wound),and study by 12 showed 21 out of 102 isolates (burn 30 (33.3%), wound 19 (15.8%),urine 29 (17.2%). A research by 13 discovered 200 individuals with burns and wounds contributed 31 staph aureus isolates. From 23 (74.19%) and 8 (25.80%) blood and burn swabs, respectively, the majority of cases were isolated. According to the findings of earlier research, the most significant sources of MDR staph aureus isolates are from the burn.
Table 3: Percentage of Antibiotics Susceptibility Profile of staph aureus Isolates Detected by DDT .
|
Antibiotics |
Susceptible (%) |
Intermediate (%) |
Resistant (%) |
|
Cefepime |
11 |
– |
89 |
|
Ceftriaxone |
20 |
– |
80 |
|
Clindamycin |
– |
– |
100 |
|
Erythromycin |
– |
– |
100 |
|
Tetracycline |
53.33 |
8.33 |
38.34 |
|
chloromphenicol |
30 |
10 |
60 |
|
ceftazidime |
10 |
– |
90 |
|
Nitrofurane |
– |
– |
100 |
Table(3), established thatstaph aureusisolates confirmed in this investigation had the highest level of resistance to most antibiotics. All of the isolates tested positive for resistance to Nitrofurane100%, which is consistent with research by a local Babylon province study by14. While study by 15 they conducted the results showed high resistance 100% to erythromycin and clindamycin, 91.6% were resistance to Ceftazidime and 81% were resistance to Tetracyclin. Medium resistance to Cefixim, Cefotaxin, Cefepim, Chloramphenicol and Ceftriaxone (73.3%, 71.6%, 68.3%, 63.3% and 56.6%), respectively. Low resistance to Ciproflaicin, Ofloxacin, Norfloxacin, Gentamicin, Piperacillin and Levofloxacin (25%, 25%, 20%, 20% and 1.6%), respectively. While the search results showed high resistance 100% to erythromycin and clindamycin , Ceftazidime but Tetracyclin, Cefixim, Cefotaxin, Cefepim, Chloramphenicol and Ceftriaxone (91%, 80%, 70%, 60%, 50%) respectively. Agree with results Alwaeli, et al., 2021 and study by 13. All isolates were subjected to tests to determine their susceptibility to 17 different antibiotics. The developed data confirmed the results, showing a high resistance rate to nitrofurane, erythromycin, clindamycine, and tetramycin and a low resistance rate to ceftazidime, cefotaxim, ceftraxone, and chloramphenicol.
Detection of Vancomycin resistance genes
The present study show that the most common vancomycin resistance genes rate in 40 isolates of staph aureus detected vanC 40(100%), VanA 20 (50%) fallowed VanB 28 (70%) as in figures (3,4,5) The results of this study showed that the isolated staph aureus have these genes at different rate. PCR study for VanC subunits of genes discovered the presence of VanA genes is 100 % among 40 staph aureus isolates, Concerning the VanB subunits of genes, The present study of primer VanB positive for 28 (70%) isolates. The amplified products as in table (1).
The result of study has been observed in Al-Diwaniya by 16 which found the most common detected VanA and VanB (100%). and study in Indonesia by 17 who found the Presence VanB (54.5%) among 11 staph aureus isolates this difference in result related to difference in the places of isolation, the community education as a source of isolates while study by Ahmed2021 found VanA 6 (37.5%)and study by18. The results of their studies indicate that 100% of staph aureus isolates harbored the VanB gene agree with our search and another study by 12. They recorded the results of VanB B gene test, the isolation rate of staph aureus was 20.5% (21/102) of samples and the positive results for VanB were found in 21 (84%)/25 tested isolates, study by 16 found. The isolates that resistance to at least one type of Vancomycin were screened for the presence of genes (VanA and VanB) by polymerase chain reaction. The gene VanB was found in 10 (100%) Vancomycin resistant isolates, followed by VanB4 (40%). Vancomycin resistance in staph aureus have overexpression efflux pump and membrane impermeability (Nouri et al.,2016). Vancomycin is used in the treatment of the following types of bacterial infections in both adult and pediatric patients . Vancomycin is a powerful antibiotic used to treat infections caused by Gram-positive bacteria, including MRSA. However, over the years, some strains of Staphylococcus aureus have developed resistance to vancomycin, rendering this antibiotic ineffective in treating these infections. Vancomycin resistance in Staphylococcus aureus occurs due to the acquisition of specific genes, often carried on plasmids or transposons, which provide resistance against the drug. The most common mechanism of resistance is the alteration of the target site of vancomycin within the bacterial cell wall. Vancomycin typically binds to the D-Ala-D-Ala terminus of the peptidoglycan precursor, preventing its incorporation into the growing cell wall and thereby inhibiting cell wall synthesis. However, in vancomycin-resistant strains, the D-Ala-D-Ala terminus is changed to D-Ala-D-Lac or D-Ala-D-Ser, reducing the binding affinity of vancomycin and allowing the bacteria to continue building their cell walls even in the presence of the antibiotic. Another mechanism of resistance involves the thickening of the bacterial cell wall, which reduces the access of vancomycin to its target site. Additionally, some MRSA strains produce enzymes that can degrade vancomycin, further diminishing its effectiveness. The emergence of vancomycin-resistant Staphylococcus aureus poses a significant challenge in healthcare settings. Patients infected with these strains have limited treatment options, often relying on alternative antibiotics that may have higher toxicity or lower efficacy 21. Moreover, the transmission of vancomycin-resistant MRSA in healthcare facilities can lead to outbreaks, making infection control measures crucial in preventing its spread. Preventing the further spread of vancomycin resistance requires a multifaceted approach. This includes prudent use of antibiotics to minimize the development of resistant strains, strict adherence to infection control protocols in healthcare settings, and ongoing research to develop new antibiotics and treatment strategies 22. Surveillance programs are also essential to monitor the prevalence of vancomycin-resistant Staphylococcus aureus and to implement timely interventions to prevent its dissemination. Combating antibiotic resistance, including vancomycin resistance, requires a multi-faceted approach involving healthcare providers, researchers, policymakers, and the public. It’s a global health challenge that necessitates a concerted effort to preserve the effectiveness of antibiotics and protect public health. Vancomycin is an antibiotic that is commonly used to treat infections caused by Gram-positive bacteria, including Staphylococcus aureus, a bacterium that can be responsible for a wide range of infections in humans. However, some strains of Staphylococcus aureus have developed resistance to vancomycin, which poses a significant challenge in the treatment of these infections 23. There are several mechanisms through which Staphylococcus aureus can develop resistance to vancomycin, and three important resistance determinants have been identified: VanA, VanB, and VanC. Unlike VanA and VanB resistance, VanC resistance is typically not transferrable between different bacterial species. VanA Resistance: VanA resistance is one of the most well-known and widespread mechanisms of vancomycin resistance in Staphylococcus aureus. It is characterized by the presence of the VanA operon, which encodes enzymes responsible for altering the structure of the bacterial cell wall, preventing vancomycin from binding to its target24. This mechanism is often associated with the transfer of resistance genes between bacteria, making it particularly problematic in the spread of resistance. The VanA resistance determinant is typically found in Enterococcus sp. but can also be transferred to Staphylococcus aureus through horizontal gene transfer. VanB resistance is another significant mechanism observed in vancomycin-resistant Staphylococcus aureus strains. Like VanA, it is associated with alterations in the bacterial cell wall that reduce vancomycin’s effectiveness.25 The VanB operon encodes enzymes that modify the peptidoglycan precursor, rendering it less susceptible to vancomycin binding. VanB resistance is typically found in Enterococcus fascism but can occasionally be transferred to Staphylococcus aureus and other Gram-positive bacteria through gene transfer mechanisms VanC Resistance:
VanC resistance, while less common in Staphylococcus aureus, is a mechanism associated with intrinsic resistance in some Gram-positive bacteria, such as Enterococcus gallinarum and Enterococcus casseliflavus. It involves changes in the structure of the bacterial cell wall precursors, which make vancomycin less effective in binding to its target.
 |
Figure 3: Showed VanC genes (150bp) ; lane L represent DNA marker size(100-500bp).Lanes (1-40 isolates) were positive results. |
 |
Figure 4: Showed VanB genes (580bp) ; lane L represent DNA marker size(100-700bp). Lanes (1-28 ) were positive while isolates NO. (29-40,) were negative |
 |
Figure 5: Showed VanA genes (520bp) ; lane L represent DNA marker size(100bp). Lanes (1- 20 ) were positive result |
Conclusion
Vancomycin resistance in Staphylococcus aureus, particularly in MRSA strains, highlights the importance of antibiotic stewardship and infection control practices. Addressing this issue requires collaborative efforts among healthcare professionals, researchers, and policymakers to preserve the effectiveness of existing antibiotics and develop innovative solutions to combat antibiotic-resistant bacteria. Vancomycin resistance in Staphylococcus aureus is a concerning issue, as it limits treatment options and can lead to severe infections. Preventing its emergence and spread through infection control and responsible antibiotic use is crucial, and ongoing research is essential to address this evolving problem in infectious diseases. Patients with VRSA infections should be the development of vancomycin resistance in Staphylococcus aureus is a concerning issue in the field of infectious diseases. It underscores the importance of responsible antibiotic use, infection control measures, and continued research into new treatment options to combat antibiotic-resistant bacteria. Addressing this problem is crucial for preserving our ability to effectively treat Staphylococcus aureus infections and safeguarding public health.managed carefully with the help of infectious disease specialists to maximize treatment outcomes
Acknowledgement
We acknowledge the College of Medicine, University of Babylon, Iraq for all facilities
Funding Sources
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
There are no conflict of nterest.
Authors’ Contribution
Hiba J. collected data, and analyzed data. Mays J. Literature reviewed, and data interpretation. Ghazi M. Ramadan statistical analysis and interpretation. Jwan A. and Hafidh l. drafted the manuscript.
Data Availability Statement
Specimens obtained from various clinical samples, among these samples are burn samples, urine samples, and samples from Blood, recovered in Hilla -hospitals.
Ethics Approval Statement
This research was granted ethical approval by both the Ethics Committee at the College of Medicine, University of Babylon, Iraq, and the Ethics Committee of the hospitals.
References
- Gardete, S., & Tomasz, A.. Mechanisms of vancomycin resistance in Staphylococcus aureus. The Journal of clinical investigation, 124(7), 2836-2840. (2014)
CrossRef - McGuinness, W. A., Malachowa, N., & DeLeo, F. R.. Focus: infectious diseases: vancomycin resistance in Staphylococcus aureus. The Yale journal of biology and medicine, 90(2), 269. (2017)
- Cong, Y., Yang, S., & Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. Journal of advanced research, 21, 169-176. (2020).
CrossRef - Wu, Q., Sabokroo, N., Wang, Y., Hashemian, M., Karamollahi, S., & Kouhsari, E. Systematic review and meta-analysis of the epidemiology of vancomycin-resistance Staphylococcus aureus isolates. Antimicrobial Resistance & Infection Control, 10, 1-13. (2021).
CrossRef - Shariati, A., Dadashi, M., Moghadam, M. T., van Belkum, A., Yaslianifard, S., & Darban-Sarokhalil, D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Scientific reports, 10(1), 12689. (2020).
CrossRef - Mahros, M. A., Abd-Elghany, S. M., & Sallam, K. I. Multidrug-, methicillin-, and vancomycin-resistant Staphylococcus aureus isolated from ready-to-eat meat sandwiches: An ongoing food and public health concern. International Journal of Food Microbiology, 346, 109165. (2021).
CrossRef - Al-Amery, K., Elhariri, M., Elsayed, A., El-Moghazy, G., Elhelw, R., El-Mahallawy, H. & Hamza, D. Vancomycin-resistant Staphylococcus aureus isolated from camel meat and slaughterhouse workers in Egypt. Antimicrobial Resistance & Infection Control, 8, 1-8. (2019).
CrossRef - Li, G., Walker, M. J., & De Oliveira, D. M. Vancomycin resistance in enterococcus and Staphylococcus aureus. Microorganisms, 11(1), 24. (2022).
CrossRef - Selim, S., Faried, O. A., Almuhayawi, M. S., Saleh, F. M., Sharaf, M., El Nahhas, N., & Warrad, M. Incidence of vancomycin-resistant Staphylococcus aureus strains among patients with urinary tract infections. Antibiotics, 11(3), 408. (2022).
CrossRef - Javed, M. U., Ijaz, M., Fatima, Z., Anjum, A. A., Aqib, A. I., Ali, M. M., … & Ghaffar, A. Frequency and Antimicrobial Susceptibility of Methicillin and Vancomycin-Resistant Staphylococcus aureus from Bovine Milk. Pakistan Veterinary Journal, 41(4). (2021).
CrossRef - Saber, T., Samir, M., El-Mekkawy, R. M., Ariny, E., El-Sayed, S. R., Enan, G. & Tartor, Y. H. Methicillin-and vancomycin-resistant staphylococcus aureus from humans and ready-to-eat meat: characterization of antimicrobial resistance and biofilm formation ability. Frontiers in Microbiology, 12, 735494. (2022).
CrossRef - Wang, S., Cai, C., Shen, Y., Sun, C., Shi, Q., Wu, N. & Zhou, H.In vitro activity of contezolid against methicillin-resistant staphylococcus aureus, vancomycin-resistant enterococcus, and strains with linezolid resistance genes from China. Frontiers in Microbiology, 12, 729900. (2021).
CrossRef - Akhtar, F., Khan, A. U., Misba, L., Akhtar, K., & Ali, A.. Antimicrobial and antibiofilm photodynamic therapy against vancomycin resistant Staphylococcus aureus (VRSA) induced infection in vitro and in vivo. European Journal of Pharmaceutics and Biopharmaceutics, 160, 65-76. (2021).
CrossRef - Shariati, A., Dadashi, M., Moghadam, M. T., van Belkum, A., Yaslianifard, S., & Darban-Sarokhalil, D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Scientific reports, 10(1), 12689. (2020).
CrossRef - ElSayed, N., Ashour, M., & Amine, A. E. K.. Vancomycin resistance among Staphylococcus aureus isolates in a rural setting, Egypt. Germs, 8(3), 134. (2018)
CrossRef - Saeed, A., Ahsan, F., Nawaz, M., Iqbal, K., Rehman, K. U., & Ijaz, T. Incidence of vancomycin resistant phenotype of the methicillin resistant Staphylococcus aureus isolated from a tertiary care hospital in Lahore. Antibiotics, 9(1), 3. (2019).
CrossRef - Guan, D., Chen, F., Xiong, L., Tang, F., Faridoon, Qiu, Y., … & Huang, W. (2018). Extra sugar on vancomycin: new analogues for combating multidrug-resistant Staphylococcus aureus and vancomycin-resistant Enterococci. Journal of Medicinal Chemistry, 61(1), 286-304.
CrossRef - Al-Amery, K., Elhariri, M., Elsayed, A., El-Moghazy, G., Elhelw, R., El-Mahallawy, H. & Hamza, D. (2019). Vancomycin-resistant Staphylococcus aureus isolated from camel meat and slaughterhouse workers in Egypt. Antimicrobial Resistance & Infection Control, 8, 1-8.
CrossRef - U C.L. Perng, T.S. Chiueh, S.Y. Lee, C.H. Chen, F.Y Chang, C.C. Wang, W.M. Chi Detection and typing of vancomycin-resistance genes of enterococci from clinical and nosocomial surveillance specimens by multiplex PCR, Epidemiol. & Infect. 126 (3), pp. 357-363(2001).
CrossRef - R. Lemcke, M. Bülte: Occurrence of the vancomycin-resistant genes vanA, vanB, vanC1, vanC2 and vanC3 in Enterococcus strains isolated from poultry and pork,Int. J. Food Microbiol., 60 (2–3) (2000), pp. 185-194
CrossRef - Cong, Y., Yang, S., & Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. Journal of advanced research, 21, 169-176. (2021).
CrossRef - Maharjan, M., Sah, A. K., Pyakurel, S., Thapa, S., Maharjan, S., Adhikari, N & Thapa Shrestha, U. Molecular confirmation of vancomycin-resistant Staphylococcus aureus with vanA gene from a hospital in Kathmandu. International Journal of Microbiology. (2021).
CrossRef - Yang, Q., Li, X., Jia, P., Giske, C., Kahlmeter, G., Turnidge, J & Chinese Committee on Antimicrobial Susceptibility Testing (ChiCAST). Determination of norvancomycin epidemiological cut-off values (ECOFFs) for Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus hominis. Journal of Antimicrobial Chemotherapy, 76(1), 152-159. (2021).
CrossRef - Shariati, A., Dadashi, M., Moghadam, M. T., van Belkum, A., Yaslianifard, S., & Darban-Sarokhalil, D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Scientific reports, 10(1), 12689. (2020).
CrossRef - Nepal, N., Mahara, P., Subedi, S., Rijal, K. R., Ghimire, P., Banjara, M. R., & Shrestha, U. T. Genotypically Confirmed Vancomycin-Resistant Staphylococcus aureus with vanB Gene Among ClinicalIsolates in Kathmandu. Microbiology Insights, 16,11786361231183675(2023)
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





