Introduction
Jack beans are one of the types of beans in Indonesia, especially in Java and Sumatra. Jack beans have nutritional contents, especially protein, which is relatively high at 24% 1. However, the use of products from Jack beans is very minimal, with only 25% of products 2. Dependence on the processing of legumes generally uses soybeans, so that the potential for Jack beans, especially protein, is not widely utilized. Products developed using Jack beans include raw materials for tempeh, milk, flour substitute for flour, cooking oil, and shredded 3,4.
Fermentation of Jack beans into tempeh produces high nutrition. The production of Tempeh starts from soaking, cooking, inoculation, storage, and packaging 5. Each processing process has a different effect on legumes’ nutritional and antinutritional components. Jack beans contain toxic substances, including choline, hydrozianine acid, trogonelin, glucocyanide, and phytic acid which are antinutritional compounds but can be neutralized and detoxified by process making tempeh 6–8. Traditional processing or a combination of these reduces antinutritional factors and increases the bioavailability of nutrients. Tempeh made of Jack beans can affect the nutrient composition that is important for health, such as increasing protein content, producing several bioactive compounds like phenolic compounds, and developing vitamins, namely vitamin B1 and vitamin B2 1,9. How an efficient process can affect the nutrient composition and the extent to which it will reduce antinutritional factors are still considered essential issues to be explored 10,11.
Soaking the beans is an essential step in the process of making tempeh, among others, facilitating the release of seeds, the absorption of water for subsequent fungal fermentation, and acid fermentation. It inhibits the growth of spoilage fungi 12. The soaking process is generally carried out using distilled water and using chlorine-free water overnight to lower the pH so that it is suitable for use mold growth and accelerated natural lactic fermentation 11,13. The soaking process has been effective in removing antinutritional factors, but some metabolic reactions affect the content of some compounds. The soaking process reduces some of the antinutritional factors and increases in vitro protein digestibility, but this is dependent on legume cultivar and soaking process conditions. The soaking process depends on the type of solution, time soaking, and temperature 6,14. Canavalia ensiformis with a ratio of water material is 1:3 in a soaking time of 72 hours to produce 36.06% crude protein 15,16. The 24 h soaking time in Canavalia ensiformis with a 1:10 ratio of materials and water resulted in 31% crude protein 17. Therefore, this is a strong assumption of the effect of the activity of microbes that can determine the nutritional value of the soaking process of Jack beans. Soaking the Jack beans is one of the pre-fermentation stages by utilizing the microorganisms that grow in them. These microorganisms have an important role in producing enzymatic hydrolytic (protease, lipase, and amylase). In the soaking process, it is found that there was the enzymatic activity of microbes produced, including Bacillus licheniformis, Bacillus pumilus, Enterococcus faecium, Pseudomonas luteola, Aerococcus viridans, Bacillus mycoides, and Staphylococcus cohnii are reported to have both protease and lipase activities 18.
Microbes have produced an enzymatic process and their ability to produce enzymes can be elucidated further using metagenomic analysis. The metagenomic analysis is a technique using a next-generation sequencing based on culture-independent microorganisms 19. Metagenomic is a precise way to determine the microbial diversity that is uncultured in an environment. Metagenomic based on DNA analysis from a community or ecosystem using the 16s rRNA gene 20. 16S rRNA marker is used for amplification DNA and identification of various diversity microbial. Therefore, to determine the microbial diversity in the soaking process of Jack beans, metagenomic analysis with next-generation sequencing is required to elucidate the effects of nutritional factors of Jack beans tempeh and the method novelty applied on soaking process Jack beans.
Materials and Methods
Sampling and Preparation
Soaked 500 grams of Jack beans from Central Java Indonesia, in sterile water with a ratio of 1:10. The soak is placed in the soaking box for 12 hours and 24 hours with 25oC and relative humidity (RH) of 60%. Ten ml of soaked water at different soaking times (12 h and 24 h) were used for the next analysis.
Metagenomic Analysis
Extraction of Bacterial Genome DNA and 16S rRNA gene amplification
Extraction of bacterial genomic DNA was carried out in water baths for 12 h and 24 h. The soaking water is filtered first using a vacuum on a 0.45 m membrane filter (Thermo Fisher Scientific, United States). The filter was extracted using the DNA Isolation Kit with the CTAB/SDS method 21,22. The extracted DNA samples were collected, centrifuged, and diluted to 1ng/μL using sterile water.
Identification of bacterial diversity 16S rRNA genes from different regions (16SV4/16SV3/16SV3-V4/16SV4-V5) were amplified using specific primers with barcodes 20,22. The PCR reactions was by Phusion® High-Fidelity PCR Master Mix (New England Biolabs). The 30 cycles of PCR (Thermo Fischer Scientific, United States) were carried out under the following conditions: pre-denaturation 94ºC for 5 minutes, denaturation 92ºC for 30 seconds, annealing 62ºC for 30 seconds, elongation 72ºC for 30 seconds, and post-elongation 72ºC for 7 minutes 23. The loading buffer was mixed once with the same volume as the PCR product and electrophoresis was operated on 2% agarose gel to detect results. DNA samples were selected with main strips between 400 bp-450 bp for further experiments. DNA bands from PCR products were cut by QIAquick Gel Extraction Kit and purified by Qiagen Gel Extraction Kit (Qiagen, Germany).
Data and Information analysis (Operational Taxonomic Unit Cluster and Alpha Diversity)
Analysis of PCR products for data and information sequences was with FLASH. FLASH is the method to combine overlapping read-paired of DNA fragments, similar DNA fragments, and a sequence of splicing. Raw tag DNA fragments from FLASH were purified and filtered to obtain high-quality clean tags by Qiime 24,25. Clean tags were compared with reference databases using the UCHIME algorithm and detected by chimera sequences. The chimera sequence can obtain an effective tag database by deleting the sequence 26. All effective tags classified with 97% similarity are considered OTU (Operational Taxonomy Units) and all sequence clones were submitted to BLASTN-NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/) 27,28. The OTUs cluster was analyzed using UPARSE software 26. The annotation of bacteria by the taxonomic tree, such as kingdom, phylum, class, order, family, genera, and species. Taxonomic trees were analyzed using Mothur software with a database from the SILVA 29,30.
The phylogenetic tree representative OTU sequences were obtained by MUSCLE software. OTU abundance information was normalized using the sequence number standard corresponding to the sample with the least sequence 31. Alpha diversity was applied in analyzing the complexity of biodiversity for the sample. The alpha diversity can indicated Observed-species, Chao1, Shannon Wiener Index, Simpson index, ACE, Good-coverage 11,32. All of these indices in our sample were calculated by QIIME and displayed with R software 24,33. All measured parameters of this study used Statistical Package for the Social Sciences Statistical software (IBM-SPSS) version 23 (NC, USA)34,35.
Results
Amplification of Gen 16S rRNA and PCR Product
The product DNA produced in each soaking was amplified using the 16S rRNA gene. The 16S rRNA amplicon is a technique microbiome analysis where different samples are analyzed simultaneously using multiplexing. The results can be used to perform microbial annotation diversity by genera, family, order, class, and phylum levels. The amplified region used is 16SV34/16S rRNA amplicon sequencing has been widely applied to the microbial community from various natural or endozoic environments such as soil, water, and host intestine. 19. The 16S rRNA gene sequence was amplified again for attachment of unique regions (multiplex or index) as a differentiator between samples, and primers for sequencing 20. The amplicons entered the cluster generation stage by MiSeq, which aimed to attach the amplicons to the flow cell and multiply the amplicons using the bridge amplification principle to form clusters. After that, the amplicons would undergo a sequencing process based on sequencing-by-synthesis (SBS) 36. The high-sequence conservation of 16S genes can separate bacteria diversity in the phylogenetic tree and identify of new taxa. Most bacterial phyla are known only from 16S surveys and have no cultured representatives 20. The electrogram amplification of Gen 16S rRNA and PCR product can be seen in Figure 1.
 |
Figure 1: 16S rRNA gene amplification and PCR Product (a) 12 h of soaking time (b) 24 h of soaking time. |
Operational Taxonomy Unit (OTUs) cluster
The number of amplicon sequences is quite large and varies because the soaking process is a treatment allowing bacteria to grow from the environment and triggers fermentation in the soaking process. The known DNA amplicons were then continued with the analysis of bacterial diversity with OTUs. A taxonomy tree was obtained could be compared with the variety of bacteria in soaking the Jack beans. OTUs analysis was done by the USEARCH algorithm and taxonomically through nucleotide blast (BLASTn) were identified 37. OTUs were created as taxonomic annotations for representative sequences of each OTUs to obtain appropriate taxa information and taxa-based abundance distributions 32,38. OTUs can analyze species annotation, species distribution, ternary plot, and phylogenetic tree. Identification and annotation of species on microbial commodities are made in clusters with 97% identity 32. Relative abundance and Heatmap OTUs of the soaking process can be seen in Figure 2.
 |
Figure 2: (a) Relative abundance and (b) Heatmap OTUs of soaking tempeh Jack Beans in different soaking times of 12 h and 24 h |
Alpha Diversity
Two samples were analyzed, namely soaking the Jack beans for 12 h and 24 h. The total sequences produced by soaking the Jack beans for 12 h produced 178762 more sequences than the soaking of the Jack beans for 24 h. A total of 3.64% of the sequences were removed after filtering the length of the sequence, the presence of ambiguous sequences, and unassigned sequences to obtain clean sequences with 163193 sequences at 12 h of soaking time and 126066 sequences at 24 h of soaking time, respectively. The soaking of Jack beans produced many sequences because the soaking process came from an environment influenced by temperature and humidity, allowing a large variety of microbes to grow 11. Total reads and clean reads sequence can be shown in Table 1.
Table 1: Alpha diversity index in different of soaking time Jack beans.
| Sample | Total reads | Clean reads | Total OTU | Shannon Wiener Index (H’) | Simpson Index (D) | Goods Coverage (%) |
| 12 h of soaking time | 178762 | 139141 | 423 | 4.231 | 0.892 | 1.000 |
| 24 h of soaking time | 163193 | 126066 | 410 | 4.262 | 0.886 | 1.000 |
Taxonomic Annotation
The taxonomic annotation on the Jack beans soaking can be seen in the phylogenetic tree in Figure 3, indicating a difference in the percentages produced in the bacterial population of 12 h soaking time compared to 24 h. The taxonomic annotation will be represented by using a phylogenetic tree. Phylogenetic analysis is useful in analyzing and illustrating gene/protein/species evolution. Besides that, phylogenetic analysis is known for an increasing number of species on DNA/genome sequenced. The phylogenetic tree will annotate the kingdom, phylum, class, order, family, genera, and species from product sample DNA extraction 32. This study suggested that the bacterial community on the soaking process was diversified, in which 423 OTUs in 12 h of soaking time and 410 OTUs in 24 h of soaking time. The major phylum of the soaking bacteria was Firmicutes and Proteobacteria, but minor phylum Bacteroidetes (Table 2). A phylogenetic tree can represent the taxonomy tree between soaking time. The results show by differentiating the number of microbial populations at 12 h of soaking time in red color and microbial populations at 24 h soaking time in blue color.
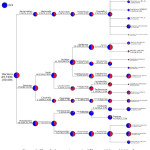 |
Figure 3: The phylogenetic tree in different 12 h and 24 h of soaking times process of Jack beans. |
Discussion
Amplification of Gen 16S rRNA and PCR Product
In the soaking process, microorganisms can grow because the appropriate temperature and humidity support them for these microorganisms 39. The foam on the surface indicated the presence of microbial growth. The soaking water was extracted using the Microbial DNA Isolation Kit to obtain DNA and amplified using the 16S rRNA gene amplification 11. Most current work with 16S rRNA produce single-end reads only up to 350 bp and paired-end reads up to 2 × 300–350 bp. The experimental diversity microbial used NGS (Next-Generation Sequencing) to estimate the taxonomic diversity of the products 19,38. The primers amplified the V1 to V9 regions of the 16S rRNA gene, resulting in an amplicon length of ~1500 bp. Species-level identification uses the first 500 bp or ~1500 bp 16S rRNA gene sequences so that they can be identified for species level in metagenomics 40,41.
The electrogram in Figure 1a shows that the 12 h of soaking time contained several colonies that were successfully amplified using the 16S rRNA sequence. Variations in amplicon length of 35 bp, 500 bp, and 5000 bp (base pair) indicated that the DNA fragment band was detected, indicating amplified bacterial fragments presence 42. Several amplicons were successfully amplified at 24 h soaking time using 16S rRNA sequences, with variations of 35 bp, 480-500bp, and 5000 bp of the amplicon, as shown in Figure 1b. At 24 h of soaking time produced different amplicons compared to 12 h of soaking time, at 480 bp-500 bp DNA amplicon produced varying thicknesses compared to 12 h of soaking time. DNA amplification in the soaking process indicated the diversity of bacteria. The amplicon of DNA at 24 h of soaking time was greater than the 12 h soaking, which would be further analyzed to determine the diversity of bacteria and taxa obtained 36.
Operational Taxonomy Unit (OTUs) cluster
The soaking process of Jack beans affects the abundance of bacteria diversity. The abundance of relative microbes can be seen in Figure 2a. The Heatmap is an interactive web with a presentation of the taxonomic annotation corresponding to OTUs in Figure 2b. This figure indicates that color coding was used to represent the percentage of each OTU to the total OTU count. The blue color contributed a low percentage of OTUs to the sample, and the red color contributed a high percentage of OTUs. In the taxa histograms, the soaking of Jack beans at 12 hours and 24 hours showed that the dominant bacteria were taxa Firmicutes, Proteobacteria and Bacteroidetes were the most important groups and accounted for 0.95, meanwhile Actinobacteria, Spirochaetes, Tenericutes, Fibrobacteres, Cyanobacteria, Nitrospirae, and Acidobacteria were also found with minor abundance. Relative species abundance refers to the biodiversity on how common one taxa is relative to other species in a particular community, especially in the process of soaking beans 43.
At 12 h soaking time of Jack beans, phylum Firmicutes, Proteobacteria Bacteroidetes value relative abundances were 0.58, 0.35, and 0.04, respectively. In 24 h of Jack bean soaking, phylum Firmicutes, Proteobacteria, and Bacteroidetes have relative abundances were 0.48, 0.44, and 0.06, respectively. The relative abundance of species and species richness in a community describes the biodiversity associated with the population of microorganisms 43. The abundance of bacteria found 89,31% of the total microorganisms in the soaking process. The abundance of the bacterial kingdom is known by analyzing the taxonomy level, such as phylum, class, order, family, genera, and species 19. Three dominant phylum’s that dominate in both processes of soaking the Jack beans are Firmicutes (52.36%), Proteobacteria (33.925%), and Bacteroidetes (3.03%).
Alpha diversity
Alpha diversity is an index of bacterial diversity that can calculate the value of Shannon-Wiener, Simpson, and good coverage index 32. Alpha diversity can determine the diversity, richness, and probability of bacteria. The alpha diversity of the soaking time index can be seen in Table 1. The soaking of Jack beans for 12 h resulted in more OTUs values than soaking at 24 h, indicating that the microbes are influenced by ideal temperature (28oC) and humidity (60%) during the 12 h soaking time. Meanwhile, at 24 h of soaking time, several dominant microbes grown can potentially have enzymatic bacteriocins, which allows these bacteria to have antimicrobial compounds. For example, Klebsiella and Lactic Acid Bacteria could act as bacteriocins 44.
The Shannon-Wiener index (H’) is a suitable index to calculate the level of species diversity 36. If H’ value is less than 1.0, the sample has low diversity and very low productivity, which indicates heavy pressure and an unstable ecosystem. If H’s value ranges from 1 < H’ > 3.5, the sample has moderate species diversity, sufficient productivity, and balanced ecosystem conditions 45. The analysis results of the two samples, namely soaking in beans for 12 h and 24 h, had a Shannon-wiener (H’) value with high species diversity, and the condition of the ecosystem species in the soaking Jack beans was quite balanced. Simpson’s index (D) is an index that calculates dominance in bacteria. If Simpson’s value is close to 0, then the value of species diversity is low 45. In the analysis results of soaking Jack beans for 12 h and 24 h, the Simpson value is more than 0, namely 0.892 and 0.882, respectively. The result showed a high species diversity, coinciding with the high value of OTUs generated in the process.
Good coverage is an estimator of the completeness of the sampling process, which is characterized by the probability that an amplicon is drawn from a random sample that has been successfully sequenced 32. Good coverage above 95% stated that the number of samples taken per sampling was sufficient to evaluate the diversity of bacteria in soaking legumes. Table 1 shows that at 12 h and 24 h beans soaking samples were successfully amplified and comprised of a range of bacterial diversity. The composition of the bacterial community in the soaking of legumes is influenced by several things, including the raw materials used, water sources, environmental conditions, and the tools used 12,45. The bacterial diversity results in the sample using NGS (Next Generation Sequencing) succeeded in identifying bacterial diversity and identified taxonomy, namely phylum, class, order, family, the genera to species 19.
Taxonomic annotation
Soaking legumes showed that the major microbial populations comprised Lactic acid bacteria, Enterobacteriaceae, and yeast. The dominant bacteria during the soaking process are caused by ambient temperature and uncontrolled conditions soaking water that make it enriched in Lactic acid bacteria 12,45. The accession number of soaking water 12 h was SAMN30222989 (https://www.ncbi.nlm.nih.gov/biosample/30222989) and soaking water 24 h was SAMN30222990 (https://www.ncbi.nlm.nih.gov/biosample/30222990).
The 100% of Kingdom Bacteria have three phyla found in soaking process 12 h and 24h. There are Firmicutes, Proteobacteria, and Bacteroidetes. The dominant phylum were Firmicutes with a relative abundance value of 53.24% (12 h of soaking time) and 47.809% (24 h of soaking time). Phylum Proteobacteria was the second population in the soaking process with 42.79% on 12 of soaking times; 38.42% on 24 h of soaking time, and the lowest relative abundance was phylum Bacteroidetes with the relative abundance of 3.97% on 12 h of soaking time; 3.567% on 24 h of soaking time.
The top 10 genera found in the soaking process of Jack beans for 12 h and 24 h of time soaking were Prevotella, Bacillus, Paenibacillus, Staphylococcus, Lactobacillus, Pediococcus, Saccharofermentants, Klebsiella, Pantoea, and Acinetobacter. The bacterial Lactobacillus casei, Streptococcus daecium, Staphylococcus epidermis, and Streptococcus dysgalactiae dominated the soaking fermentation process of tempeh12. Some other species such as Klebsiella pneumoniae, Klebsiella ozaenae, Enterobacter cloacae, Enterobacter agglomerans, Citrobacter diversus and Bacillus brevis, and Bacillus pumilus found in soaking process of tempeh 46. Phylum Firmicutes were dominant in the soaking process of Jack beans. It consists of 2 major classes which were ±50% for Bacilli and ±1.3% for Clostridia. The class of Bacilli found three families were Bacillaceae, Paenibacillaceae, and Staphylococcaceae. Two OTUs of the genera Bacillus were detected in this soaking process for 12 h and 24 h; Bacillus drentensis and Bacillus gibsonii. The soaking process of peanut seeds found Bacillus spp, which significantly affected crop yields 47. Bacillus spp. is a soil bacterium that is often found in the rhizosphere of plants. Some species produce extracellular enzymes that can hydrolyze complex proteins and polysaccharides. As the most significant bacteria, Bacillus produces an enzyme in large quantities 48. Bacillus drentensis is one bacterium can produce an enzyme, such as amylase 49. These bacteria are known for their ability to secrete large amounts of alkaline proteases with significant proteolytic activity and it is stable at considerably high pH, temperatures, and saline environments 50. Bacillus is a Gram-positive bacterium, aerobic-facultative endospore-forming motile bacteria that belongs to the family Bacillaceae. Bacillus gibsonii is one bacterium from the soil that significantly enhanced beans and wheat growth. Bacillus gibsonii is a novel protease producer, which is more closely related to the high-alkaline proteases belonging to the Subtilisin family 51.
In the soaking of Jack beans, Paenibacillus and Lactobacillus species were the most dominant group in the soaking process. Two major species of Paenibacillus glycanilyticus and Paenabacillus xylanilyticus were identified. Paenibacillus is one of the endophytic bacterium found in peanut seeds with several special properties, such as the formation of endospores, cell motility, tolerance to high osmotic pressure, and amylase activity 52. Paenibacillus glycanilyticus is a novel species to degrade heteropolysaccharides and produce catalase 53. Paenibacilli can metabolize aliphatic and aromatic organic pollutants by applying oxygenase’s, dehydrogenases, and ligninolytic enzymes 54. Based on Figure 3, Staphylococcus sp is one of the genera identified in soaking Jack beans for 12 h and 24 h where Staphylococcus warneri was identified. In the soaking process of various types of beans, the growth of Staphylococcus was found when the pH was increased, but its growth was inhibited due to the presence of Lactobacillus plantarum bacteria 55. S. warneri is a new source of lipase, extracellular protease, and the proteolytic cascade. In addition, S. warneri is a bacterium that can be an antimicrobial agent, namely bacteriocin 56.
On Lactobacillus genera, Lactobacillus fermentum species dominated at 12 h of soaking time with a percentage of 0.003%; however, the bacterium was not found at 24 h of soaking time. Pediococcus genera was dominated at 12 h of soaking time with a percentage of 1.15%, while in 24 h of soaking time with a percentage of 1.032%. In the species, Pediococcus acidilactici was found to be dominant at 12 h of soaking time. On family Lactobacillaceae, two genera were found in the soaking process. There are Lactobacillus and Pediococcus, with each species were Lactobacillus fermentum and Pediococcus acidilactici. L. fermentum is one of the Lactic acid bacteria capable of exhibiting amylolytic activity, generally isolated from soil 57. L. fermentum is a starter culture that could optimize fermented African kale’s nutrient components. L. fermentum can hydrolyze proteins, so it can form unique fermented flavours and an important proteolytic enzyme, with protease activity of 20 U/ml 58. P. acidilactici is a probiotic bacterium with high antioxidant, cryoprotective activity against alloxan, and produced extracellular proteolytic 59.
The Class Clostridia found in process soaking in 12 h and 24 h with lower relative abundance was 1.42% dan 1.280%, respectively. One OTUs genera found in the soaking process was Saccharofermentans. Saccharofermentans is Gram-positive bacterium, non-motile, and aerobic growth. Saccharofermentans is predominant cellular fatty acids, and several hexose, polysaccharides, and alcohols are fermented with fermentation products from glucose 60. The Bacteroidetes had a minor abundance phylum in the soaking process, approximately 3% of total taxa. In this study, the only genera found was Prevotella with six OTUs species. The Six OTUs species include Protovella ruminicola, Protovella sp, bacterium Protovella XPB, uncultured Protovella sp, uncultured bacterium Protovella, and unidentified bacterium Protovella. Protovella are the important bacteria that play important role in the metabolism of protein with proteolytic and characteristic of dipeptidyl peptidase and produce degradative enzymes, such as amylase and xylanase 61–63.
Phylum Proteobacteria is the second dominant population of bacteria in the soaking process. One class bacterium found in the process was Gammaproteobacteria with two Orders such as Enterobacteriales and Pseudomonadales. At 12 h soaking time, the relative abundance value of phylum proteobacteria was lower than 24 h of soaking time. Five OTUs genera found in the process soaking includes Klebsiella aerogenes, Pantoea, Acinetobacter baumannii, Acinetobacter calcoaceticus, and Acinetobacter johnsonii. Proteobacteria are classified as pathogenic bacteria, but these bacteria can produce health benefits in the tempeh process. Klebsiella is one type of bacteria that can synthesize vitamin B12, a resource for vegetarian’s. Klebsiella has a role bacterium that can produce acidification (decrease until pH 4) during the soaking process 64. Meanwhile, Klebsiella has the potential to produce bacteriocins and hydrolytic enzymes 44. Acinetobacter is a group bacterium commonly found in the environment, mostly water. Acinetobacter baumannii is a Gram-negative and amylolytic bacterium, and a potential proteolytic activity 65.
Conclusion
Soaking is an essential step in making tempeh a pre-fermentation process utilizing microbial enzymes to increase product nutrition. Soaking has uncultured microbial diversity analyzed using metagenomic data with next-generation sequencing. The different times of soaking for 12 h and 24 h gave a different percentage of microbial population, indicate that the uniqueness of fermentation of Jack beans tempeh. Firmicutes phylum was the predominant phylum in both times soaking processes. Ten OTUs genera were found in both soaking times; Prevotella, Bacillus, Paenibacillus, Staphylococcus, Lactobacillus, Pediococcus, Saccharofermentants, Klebsiella, Pantoea, and Acinetobacter. Six OTUs genera bacteria were dominant in soaking for 12 h, and four OTUs genera were dominant in soaking for 24 h. The microbial diversity indicated different enzymes were presented and contributed nutrients Jack beans Tempeh.
Acknowledgement
The authors would like to thank Universitas Padjadjaran and the Ministry of Education and Culture of Indonesia supporting our research and PT. Genetika Science Indonesia for supporting our analysis.
Conflict of Interest
All authors have no conflict of interest.
Funding Sources
This study was supported by Riset Disertasi Doktor Universitas Padjadjaran (RDDU) research scheme by Number Grant (No: 1932/UN6.N/LT/2019) from Ministry of Education, Culture, Research and Technology, Indonesia.
References
- Widaningrum N, Sukasih E, Purwani EY. Introductory Study On Processing Of Fermented Jack Bean (Canavalia Ensiformis). J Penelit Pascapanen Pertan. 2017;12(3):129. doi:10.21082/jpasca.v12n3.2015.129-136
CrossRef - Sumberdana I, Sumba I, Ustriyana I. Inventory Analysis of Raw Materials Jack Beans (Canavalia ensiformis) On UD Arjuna Bali, Nyanglan Village, District Banjarangkan, Klungkung. J Agribus Agritourism. 2016;4(5):345-354.
CrossRef - Andriati N, Anggrahini S, Setyaningsih W, Sofiana I, Pusparasi DA, Mossberg F. Physicochemical characterization of jack bean (Canavalia ensiformis) tempeh. Food Res. 2018;2(5):481-485. doi:10.26656/fr.2017.2(5).300
CrossRef - Ningrum A, Anggrahini S, Setyaningsih W. Valorization of Jack Bean as Raw Material for Indonesian Traditional Food Tempeh and Its Functional Properties. J Appl Sci. 2019;19(2):56-61. doi:10.3923/jas.2019.56.61
CrossRef - Cao ZH, Green-Johnson JM, Buckley ND, Lin QY. Bioactivity of soy-based fermented foods: A review. Biotechnol Adv. 2019;37(1):223-238. doi:10.1016/j.biotechadv.2018.12.001
CrossRef - El-Adawy TA, Rahma EH, El-Bedawy AA, Sobihah TY. Effect of soaking process on nutritional quality and protein solubility of some legume seeds. Nahrung – Food. 2000;44(5):339-343. doi:10.1002/1521-3803(20001001)44:5<339::AID-FOOD339>3.0.CO;2-T
CrossRef - Widiantara T, Kastaman R, Setiasih IS, Muhaemin M. Reduction Model Of Cyanide and Protein Content on the Jackbeans Using CMS Method ( Circulation Mixing System ). Int Conf “Food A Good Life” Jakarta, Indones. 2016;(May).
- Puspitojati E, Indrati R, Cahyanto MN, Marsono Y. Jack bean as tempe ingredients: The safety study and fate of protein against gastrointestinal enzymes. IOP Conf Ser Earth Environ Sci. 2019;346(1):0-8. doi:10.1088/1755-1315/346/1/012070
CrossRef - Affandi DR, lshartani D, Wijaya K. Physical, chemical and sensory characteristics of jack bean (Canavalia ensiformis) tempeh flour at various drying temperature. 3Rd Int Conf Condens Matter Appl Phys. 2020;2220:070004. doi:10.1063/5.0004674
CrossRef - Ragab HI, Kijora C, Abdel Ati KA, Danier J. Effect of traditional processing on the nutritional value of some legumes seeds produced in Sudan for poultry feeding. Int J Poult Sci. 2010;9(2):198-204. doi:10.3923/ijps.2010.198.204
CrossRef - Radita R, Suwanto A, Kurosawa N, Wahyudi AT, Rusmana I. Metagenome analysis of tempeh production: Where did the bacterial community in tempeh come from? Malays J Microbiol. 2017;13(4):280-288. http://www.ncbi.nlm.nih.gov/BLAST/
CrossRef - Mulyowidarso RK, Fleet GH, Buckle KA. The microbial ecology of soybean soaking for tempe production. Int J Food Microbiol. 1989;8(1):35-46. doi:10.1016/0168-1605(89)90078-0
CrossRef - Yan Y, Wolkers-Rooijackers J, Nout MJR, Han B. Microbial diversity and dynamics of microbial communities during back-slop soaking of soybeans as determined by PCR-DGGE and molecular cloning. World J Microbiol Biotechnol. 2013;29(10):1969-1974. doi:10.1007/s11274-013-1349-6
CrossRef - Kajihausa OE, Fasasi RA, Atolagbe YM. Effect of Different Soaking Time and Boiling on the Proximate Composition and Functional Properties of Sprouted Sesame Seed Flour. Niger Food J. 2014;32(2):8-15. doi:10.1016/s0189-7241(15)30112-0
CrossRef - Nishizawa K, Arii Y. Reversible changes of canavalin solubility controlled by divalent cation concentration in crude sword bean extract. Biosci Biotechnol Biochem. 2016;80(12):2459-2466. doi:10.1080/09168451.2016.1224642
CrossRef - Michael K, Sogbesan O, Onyia L. Effect of Processing Methods on the Nutritional Value of Canavalia ensiformis Jack Bean Seed Meal. J Food Process Technol. 2018;9(12):1-5. doi:10.4172/2157-7110.1000766
CrossRef - Doss A, Pugalenthi M, Vadivel VG, Subhashini G, Anitha Subash R. Effects of processing technique on the nutritional composition and antinutrients content of under-utilized food legume Canavalia ensiformis L.DC. Int Food Res J. 2011;18(3):965-970.
- Berber D, Birbir M. Determination of Major Problems of Raw Hide and Soaking Process in Leather Industry. Int J Adv Eng Pure Sci. 2019;(1):118-125. doi:10.7240/jeps.470865
CrossRef - Shah SHJ, Malik AH, Zhang B, Bao Y, Qazi J. Metagenomic analysis of relative abundance and diversity of bacterial microbiota in Bemisia tabaci infesting cotton crop in Pakistan. Infect Genet Evol. 2020;84:104381. doi:10.1016/j.meegid.2020.104381
CrossRef - Imchen M, Kumavath R, Vaz ABM, et al. 16S rRNA Gene Amplicon Based Metagenomic Signatures of Rhizobiome Community in Rice Field During Various Growth Stages. Front Microbiol. 2019;10(September):1-15. doi:10.3389/fmicb.2019.02103
CrossRef - Efriwati, Suwanto A, Rahayu G, Nuraida L. Population Dynamics of Yeasts and Lactic Acid Bacteria (LAB) During Tempeh Production. HAYATI J Biosci. 2013;20(2):57-64. doi:10.4308/hjb.20.2.57
CrossRef - Sambrook R. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory; 2000. doi:10.1101/pdb.prot100461
CrossRef - Barus T, Suwanto A, Agustina W. Metagenomic Analysis of Bacterial Diversity in Tempe using Terminal Restriction Fragment Length Polymorphism (T-RFLP) Technique. 2010;15(April):2.
CrossRef - Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rrna gene sequences from microbial communities. Curr Protoc Bioinforma. 2011;(SUPPL.36). doi:10.1002/0471250953.bi1007s36
CrossRef - Ramírez-Guzmán A, Taran Y, Armienta MA. Geochemistry and origin of high-pH thermal springs in the Pacific coast of Guerrero, Mexico. Geofis Int. 2004;43(3):415-425. doi:10.1038/nmeth.f.303.QIIME
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194-2200. doi:10.1093/bioinformatics/btr381
CrossRef - McGinnis S, Madden TL. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(WEB SERVER ISS.):20-25. doi:10.1093/nar/gkh435
CrossRef - Torkian B, Hann S, Preisner E, Norman RS. BLAST-QC: Automated analysis of BLAST results. Environ Microbiomes. 2020;15(1):1-8. doi:10.1186/s40793-020-00361-y
CrossRef - Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):590-596. doi:10.1093/nar/gks1219
CrossRef - Pruesse E, Quast C, Knittel K, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188-7196. doi:10.1093/nar/gkm864
CrossRef - Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792-1797. doi:10.1093/nar/gkh340
CrossRef - Pangastuti A, Alfisah RK, Istiana NI, et al. Metagenomic analysis of microbial community in over-fermented tempeh. Biodiversitas. 2019;20(4):1106-1114. doi:10.13057/biodiv/d200423
CrossRef - Fox J, Leanage A. R and the Journal of Statistical Software. J Stat Softw. 2016;73(2). doi:10.18637/jss.v073.i02
CrossRef - Landau S, Everitt B. A Handbook of Statistical Analyses Using SPSS. Champan Hall CRC Press; 2004.
CrossRef - Rahman A, Muktadir G. SPSS : An Imperative Quantitative Data Analysis Tool for Social Science Research. 2021;(October). doi:10.47772/IJRISS.2021.51012
CrossRef - Bukin YS, Galachyants YP, Morozov I V., Bukin S V., Zakharenko AS, Zemskaya TI. The effect of 16s rRNA region choice on bacterial community metabarcoding results. Sci Data. 2019;6:1-14. doi:10.1038/sdata.2019.7
CrossRef - Tonge DP, Pashley CH, Gant TW. Amplicon -based metagenomic analysis of mixed fungal samples using proton release amplicon sequencing. PLoS One. 2014;9(4). doi:10.1371/journal.pone.0093849
CrossRef - Sassoubre LM, Yamahara KM, Boehm AB. Temporal stability of the microbial community in sewage-polluted seawater exposed to natural sunlight cycles and marine microbiota. Appl Environ Microbiol. 2015;81(6):2107-2116. doi:10.1128/AEM.03950-14
CrossRef - Nurdini AL, Nuraida L, Suwanto A, Suliantari. Microbial growth dynamics during tempe fermentation in two different home industries. Int Food Res J. 2015;22(4):1668-1674.
- Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic Classification of Bacterial 16S rRNA Genes Using Short Sequencing Reads: Evaluation of Effective Study Designs. PLoS One. 2013;8(1):18-23. doi:10.1371/journal.pone.0053608
CrossRef - Sune D, Rydberg H, Augustinsson ÅN, Serrander L, Jungeström MB. Optimization of 16S rRNA gene analysis for use in the diagnostic clinical microbiology service. J Microbiol Methods. 2020;170(January):105854. doi:10.1016/j.mimet.2020.105854
CrossRef - Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2):330-339. doi:10.1016/j.mimet.2007.02.005
CrossRef - Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013;7(6):1092-1101. doi:10.1038/ismej.2013.10
CrossRef - Gundogan N. Klebsiella. Encycl Food Microbiol Second Ed. 2014;2:383-388. doi:10.1016/B978-0-12-384730-0.00172-5
CrossRef - Radita R, Suwanto A, Kurosawa N, Wahyudi AT, Rusmana I. Firmicutes is the predominant bacteria in tempeh. Int Food Res J. 2018;25(6):2313-2320.
- Adeniran O, Atanda O, Edema M, Oyewol O. Effect of Lactic Acid Bacteria and Yeast Starter Cultures on the Soaking Time and Quality of “Ofada” Rice. Food Nutr Sci. 2012;03(02):207-211. doi:10.4236/fns.2012.32030
CrossRef - Pandango S, Widnyana IK, Sapanca PLY. The Effect of Seed Soaking Time with Bacillus SPP Bacteria on Growth and Products of Plant Peanut (Arachis hypogaea L). Agrimeta. 2018;08(16):50-55.
- Danilova I, Sharipova M. The practical potential of bacilli and their enzymes for industrial production. Front Microbiol. 2020;11(August). doi:10.3389/fmicb.2020.01782
CrossRef - Manohar P, Sivashanmugam K. A novel α -amylase producing Bacillus drentensis isolated from coastal mud samples. 2016;(December).
- Sudha J, Ramakrishnan V, Madhusudhanan N, Mandal AB, Gurunathan T. Studies on Industrially Significant Haloalkaline Protease from Bacillus sp. JSGT Isolated from Decaying Skin of Tannery. J Adv Lab Res Biol. 2010;1(1):46-51. https://e-journal.sospublication.co.in/index.php/jalrb/article/view/13%0Ainternal-pdf://0.0.5.108/13.html
- Deng A, Zhang G, Shi N, Wu J, Lu F, Wen T. Secretory expression, functional characterization, and molecular genetic analysis of novel halo-solvent-tolerant protease from Bacillus gibsonii. J Microbiol Biotechnol. 2014;24(2):197-208. doi:10.4014/jmb.1308.08094
CrossRef - Li L, Zhang Z, Pan S, Li L, Li X. Characterization and Metabolism Effect of Seed Endophytic Bacteria Associated With Peanut Grown in South China. Front Microbiol. 2019;10(November):1-11. doi:10.3389/fmicb.2019.02659
CrossRef - Dasman A, Kajiyama S, Kawasaki H, et al. Paenibacillus glycanilyticus sp. nov., a novel species that degrades heteropolysaccharide produced by the cyanobacterium Nostoc commune. Int J Syst Evol Microbiol. 2002;52(5):1669-1674. doi:10.1099/ijs.0.02171-0
CrossRef - Rütering MP. Exopolysaccharides by Paenibacilli : from Genetic Strain Engineering to Industrial Application. Published online 2019.
- Ashenafi M, Busse M. Growth of Staphylococcus aureus in fermenting tempeh made from various beans and its inhibition by Lactobacillus plantarum. Int J Food Sci Technol. 1992;27(1):81-86. doi:10.1111/j.1365-2621.1992.tb01182.x
CrossRef - Héchard Y, Ferraz S, Bruneteau E, Steinert M, Berjeaud JM. Isolation and characterization of a Staphylococcus warneri strain producing an anti-Legionella peptide. FEMS Microbiol Lett. 2005;252(1):19-23. doi:10.1016/j.femsle.2005.03.046
CrossRef - Padmavathi T, Bhargavi R, Priyanka PR, Niranjan NR, Pavitra PV. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J Genet Eng Biotechnol. 2018;16(2):357-362. doi:10.1016/j.jgeb.2018.03.005
CrossRef - Sun F, Hu Y, Yin X, Kong B, Qin L. Production, purification and biochemical characterization of the microbial protease produced by Lactobacillus fermentum R6 isolated from Harbin dry sausages. Process Biochem. 2020;89:37-45. doi:10.1016/j.procbio.2019.10.029
CrossRef - Song YR, Lee CM, Lee SH, Baik SH. Evaluation of probiotic properties of pediococcus acidilactici m76 producing functional exopolysaccharides and its lactic acid fermentation of black raspberry extract. Microorganisms. 2021;9(7). doi:10.3390/microorganisms9071364
CrossRef - Chen, Liu, Zhang. Saccharofermentans. Bergey’s Man Syst Archaea Bact. Published online 2017:1-5. doi:10.1002/9781118960608.gbm01450
CrossRef - Griswold KE, White BA, Mackie RI. Diversity of extracellular proteolytic activities among Prevotella species from the rumen. Curr Microbiol. 1999;39(4):187-194. doi:10.1007/s002849900443
CrossRef - Okabe S, Shimazu Y. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: Effects of temperature and salinity. Appl Microbiol Biotechnol. 2007;76(4):935-944. doi:10.1007/s00253-007-1048-z
CrossRef - Flint HJ, Duncan SH. Bacteroides and Prevotella. Vol 1. Second Edi. Elsevier; 2014. doi:10.1016/B978-0-12-384730-0.00031-8
CrossRef - Barus T, Hanjaya I, Sadeli J, Lay BW, Suwanto A, Yulandi A. Genetic Diversity of Klebsiella spp. Isolated from Tempe based on Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction (ERIC-PCR). HAYATI J Biosci. 2013;20(4):171-176. doi:10.4308/hjb.20.4.171
CrossRef - Amin M, Yusuf MF, Suarsini E, et al. Identification of indigene bacteria from waste water of Regional Public Hospitals in Pacitan. IOP Conf Ser Earth Environ Sci. 2019;230(1). doi:10.1088/1755-1315/230/1/012089.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.







