Introduction
With the aquaculture industry’s rapid development, the probability of fish serving as a vector for human enteropathogenic bacteria has received increased attention 1. Enteric and other infectious diseases have been linked to the existence of certain bacterial species such as Listeria, Aeromonas, Pseudomonas, Staphylococcus, Salmonella and Escherichia, on the flesh of fish, indicating that they have the ability to cause human disease when consumed or handled 2-7. Gram-negative bacteria such as Aeromonas hydrophila, Citrobacter freundii, Escherichia coli, Enterobacter aerogenes, Vibrio anguillarum, Klebsiella, and Pseudomonas and Gram-positive bacteria such as Listeria, Bacillus and Staphylococcus were present in abundance in the fish’s intestine 1,8. These bacteria exist with high levels in the different organs of the fish intestines (i.e. liver, kidney, intestine etc.) which may be a great source of cross-contamination during processing.
Antibiotic-resistant E. coli derived from animals with elevated human carriage rates should not be ignored, as according to evidence, they contribute to the burden of human microbial resistance 9,10.
The fish farming industry is rapidly expanding, particularly in Asia (including in Vietnam) 11. Besides, Vietnam’s main source of foreign exchange is fisheries exports, aquaculture production accounts for about 65% of Vietnam’s total fisheries exports. Pangasius (catfish) is the fisheries sector’s second-largest commodity (behind shrimp), accounting for about half of the total export value 12. The use of antibiotics extensively in aquaculture has resulted in the selection of multi-resistant bacteria in the fish gut flora, which are then transferred to the human gut commensal flora when the fish are consumed. The widespread use of anti-microbials such as oxolinic acid, tetracycline, florfenicol, and nitrofurantoin has resulted in the selection of resistance in fish pathogens 11,13.
In view of these facts, this study was to establish a baseline understanding for the existence of antibiotic-resistant E. coli in starved and non-starved Pangasius viscera. This aimed to assess the possible resistance to antibiotics and cross-contamination of the bacterial pathogens into the muscles of the fish during processing, as well as to evaluate the effect of starving Pangasius fish to limit cross-contamination in the products at the processing companies in the Mekong Delta, Vietnam.
Material and Methods
Sampling
Thirty of Pangasius viscera samples including 14 samples of starved fish and 16 samples of non-starved fish were collected randomly from two Pangasius processing company in An Giang and Dong Thap provinces, Vietnam from November 2018 to July 2020.
Samples of the whole Pangasius fish weighing approximately 2-3 kg were put into sterile bags, sealed and transported to Can Tho University’s Food Microbiology Laboratory in insulated ice boxes within 2-3 hours after collection for analyzing bacterial criteria.
Microbial analysis
The whole of the viscera of raw Pangasius fish were taken and transferred aseptically to new sterile containers (Stomacher bags, France) via sterile scalpels and tweezers. Twenty-five grams of these viscera samples taken from different parts (e.g. intestine, liver, kidney, stomach etc.) were transferred aseptically to a stomacher bag using sterile scissors and tweezers. Maximum Recovery Diluent (MRD, Merck, Darmstadt, Germany) was used for a ten-fold dilution, then the samples mixture was homogenized for five minutes. Subsequently, a decimal dilution, using 1 mL of the samples was made in MRD of 9 mL. Total aerobic mesophilic counts (TMC) and total anaerobic mesophilic counts (TAMC) was counted by pour-plating 1 mL of appropriate sample dilutions on Plate Count Agar (PCA, Merck, Darmstadt, Germany), with an over-layer for TAMC, followed by incubation at 37oC for 48-72 h. E. coli and Coliforms were counted by the spreading of 0.1 mL of appropriate sample dilutions on Coliform Agar Enhanced Selectivity (Coliform Agar ES, Merck, Darmstadt, Germany) and incubated at 37oC for 24 h (blue to violet colonies were counted as E. coli and pink to red colonies were counted as Coliforms-including E. coli group). For the determination of mesophilic lactic acid bacteria (LAB), de Man Rogosa Sharpe agar media (MRS, Merck, Darmstadt, Germany) was used by pour-plating with an over-layer and then incubation for 48-72 h at 37°C. After incubation, the bacterial colonies were counted manually and presented as logarithms (log CFU g-1).
For E. coli, after counting on Coliform Agar ES, three to five colonies (or fewer if five were not available or showed confluent growth) of each observed morphology (color, margin, surface and shape) were selected and then isolated to collect the pure colonies and carry out biochemical tests.
The confirmation of E. coli colonies was done using five biochemical tests: Indole, Methyl red, Voges-Proskauer, Citrate and Kligler Iron Agar test (Merck, Darmstadt, Germany) 14. All the confirmed E. coli isolates were then stored under -80oC in order to perform antibiotic susceptibility testing.
Antibiotic resistance test
The disk diffusion method was used to test antibiotic susceptibility of the E. coli isolates using Mueller-Hinton agar plates (MHA, Merck, Darmstadt, Germany) 15. As a control sample, E. coli strain ATCC 25922 was used. Antimicrobial agents used 14 were: ampicillin 10 µg (AMP), meropenem 10 µg (MER), gentamicin 10 µg (GEN), tetracycline 30 µg (TET), chloramphenicol 30 µg (CHL), ciprofloxacin 5 µg (CPR), fosfomycin 200 µg (FOS) (Abtek, United Kingdom), ceftazidime 30 µg (Cz), cefotaxime 30 µg (Ct), cefoxitin 30 µg (Cn), kanamycin 30 µg (Kn), streptomycin 10 µg (Sm), sulfamethoxazole/trimethoprim 23.75/1.25 µg (Bt), nalidixic acid 30 µg (Ng) and colistin 10 µg (Co) (Nam Khoa, Vietnam). The E. coli isolates were labeled as susceptible, intermediate, and resistant to the antibiotics according to the zone diameter interpretative standards recommended by Clinical and Laboratory Standards Institute (2019) 15.
Statistical analysis
Microsoft Excel version 2019 (Microsoft Office, U.S.A.) was used to compute and graph the data. The microbial counts were compared using analysis of variance with significance level of 5% via SPSS Statistics version 20.0 (SPSS Inc., Chicago, U.S.A.). The data were reported as the mean value ± standard deviation.
Results and Discussion
Microbial counts of the Pangasius viscera
Figure 1 shows microbial loads (i.e. total counts of TMC and TAMC), LAB, coliforms and E. coli of non-starved and starved Pangasius viscera. Specifically, TMC, TAMC, LAB, coliforms and E. coli on viscera of non-starved Pangasius were 7.9±0.3, 7.9±0.4, 7.0±0.5, 5.5±0.9 and 5.4±1.0 log CFU g-1, respectively. TMC, TAMC, LAB, coliforms and E. coli on viscera of starved Pangasius were 6.7±1.2, 6.8±1.0, 2.6±0.8, 3.8±0.4 and 3.1±0.3 log CFU g-1, respectively (Fig. 1). These results showed that no significant difference was found in the total anaerobic counts (p = 0.179) as well as the total aerobic counts (p = 0.160) on the viscera. There were significant differences found in LAB, coliforms and E. coli between non-starved and starved Pangasius viscera (p = 0.018, p = 0.006 and p = 0.000, respectively).
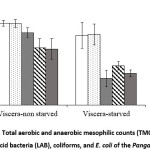 |
Figure 1: Total aerobic and anaerobic mesophilic counts (TMC and TAMC), lactic acid bacteria (LAB), coliforms, and E. coli of the Pangasius viscera |
The obtained results confirmed that starving Pangasius fish before slaughtering was effective at significantly reducing the intestinal microflora. The bacterial counts in digestive tract of fish can reach up to 8 log CFU g-1. The number of microorganisms depended on various factors including the seasons, part of the digestive tracts of fish, and feeding types 16. Depending on environmental conditions, the common microbiota in fish’s gastrointestinal tract include Vibrio, Aeromonas, Flavobacterium, Plesiomonas, Pseudomonas, Enterobacteriaceae, Micrococcus, Acinetobacter, Clostridium, Fusariumand, Bacteroides which may vary from species to species 1,17. Diet (i.e. starvation) plays a major factor in forming gut microbiota and diverse bacterial species in the gut microbiota were extremely responsive to starvation 18. In this study, the pathogens i.e. coliforms and E. coli enumerated on the viscera of starved Pangasius decreased significantly compared to the non-starved Pangasius. Therefore, starving of Pangasius fish before slaughtering (from two to three days) would be effective in partly limiting cross-contamination especially pathogenic and spoilage bacteria (i.e. LAB) for fish fillets during processing due to widely distributed microorganisms in the intestinal tracts 19. The previous studies also reported that endogenous bacteria of intestinal tracts can contaminate at the filleting step due to viscera perforation 4,20.
Antibiotic resistance
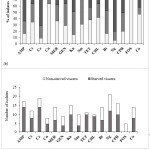
Fig. 2-a showed the high level of ampicillin resistance in E. coli isolates (63.64%), ceftazidime (69.09%) and nalidixic acid (78.18%), followed by sulfamethoxazole/trimethoprim, ciprofloxacin and kanamycin (45.5-49.1%); and chloramphenicol, cefotaxime, meropenem, tetracycline and colistin (36.4-41.8%). In contrast, the isolates were mostly susceptible to fosfomycin (83.64%). Fig. 2-b showed the prevalence of antibiotic-resistant E. coli isolated from the viscera of non-starved and starved Pangasius. The incidence rate of resistant isolates obtained from the viscera of non-starved Pangasius (47.8-71.2%) was seemingly higher compared to that from the starved Pangasius (16.7-52.2%). There were 38/55 (69.09%) isolates of multiple resistance E. coli (resistance to three or more kinds of antibiotics) among the samples. Surprisingly, 5/55 (9.09%) isolates were resistant to 15 antibiotics tested (Fig. 3).
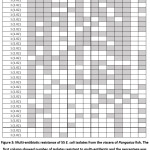 |
Figure 3: Multi-antibiotic resistance of 55 E. coli isolates from the viscera of Pangasius fish. |
A study of Salako et al. 21 reported that 61-69% of E. coli isolated from Pangasius freezing process at two factories in Mekong Delta Vietnam were resistant to ampicillin (43-47%), followed by cefotaxime (33-40%); and, prevalence of multi-drug resistance of the isolated E. coli was also reflected. It may explain that the accumulation of antibiotics in the water, environment, and ponds resulted in the occurrence of antibiotic resistant bacteria isolated from Pangasius fish during farming. Generally, starving of Pangasius fish before slaughtering would be effective in partly limiting the cross-contamination of fish fillet with pathogenic, spoilage, and antibiotic resistant bacteria during processing.
There was a connection between antibiotic use and the development of antibiotic resistance in bacterial pathogens 22. It was recorded that there were more than 70 notifications of frozen Pangasius products contaminated with antibiotic substances i.e. chloramphenicol, ciprofloxacin, enrofloxacin, neomycin, etc. 23 which showed an evident of uncontrolling well the use of antibiotics during Pangasius fish farming. On the other hand, the prevalence of multi-antibiotic resistance of bacteria in pond water of cultured fish and fresh Pangasius fish has also been reported 24,25. Jiang et al. 11 also reported that high levels of resistance to ampicillin, florfenicol, tetracycline and co-trimoxazole were found in 218 E. coli isolates recovered from farmed fish gut. In addition, some studies have shown that antibiotic use in animal husbandry can contribute significantly to the selection and spread of antibiotic-resistant bacteria in the environment 26,27. Furthermore, aquacultural products are sometimes at risk of antibiotic-resistant bacteria through the food chain and from handlers 28,29. In contrast, a number of studies have mentioned that antibiotic overuse did not always result in an increase in resistance because of a complex relationship between the use of antibiotics and the occurrence of antibiotic-resistant bacteria 30,31; antibiotic-resistant bacteria might be abundant in the environment even when the corresponding antibiotics are not available and in the presence of co-selection by other antibiotics, horizontal gene transfer of resistant genes can play a key role in their dissemination in the environment 32,33.
The emergence of E. coli isolates with multiple antibiotic-resistant phenotypes (more detail in Fig. 3), involving co-resistance to four or more different antibiotic families has been previously mentioned and is regarded a serious health concern 34,35. As E. coli is a potential vector for antibiotic resistance gene transfer, the possibility of horizontal transfer to human pathogens may occur 22. In the present study, the origin of antibiotic resistance bacteria can be determined from viscera of Pangasius where they can contaminate into Pangasius fillets during processing. Hence, it should be taken into account that the cross-contamination of bacteria, especially antibiotic resistant bacteria can be limited during processing steps if the bursting of gut is avoided during processing i.e. gutting and filleting. This study, as far as we know, is one of the first report highlighting the microbial loads and the incidence of antibiotic-resistant E. coli isolates derived from the viscera of Pangasius. Intervention to control antimicrobial resistant bacteria during farming and processing is necessary in the follow-up studies.
Conclusions
Generally, lower counts of lactic acid bacteria, coliform and E. coli in viscera of starved Pangasius compared to that of non-starved Pangasius was observed. The results of E. coli isolated from the Pangasius viscera showed that there were 69.09% of the isolates which were multi-antibiotic resistant. It was determined that there was a high level of resistance to ampicillin, ceftazidime and nalidixic acid. A total of 9.09% of isolates were resistant to fifteen antibiotics tested. There is a need for both the prudent use of these antimicrobial agents in aquaculture and stringent appropriate infection control in Pangasius processing chain in industry. Besides, treatments to control effectively antimicrobial resistant bacteria are also suggested for study in subsequent research.
Acknowledgment
We would also like to thank the companies involved and allowed to publish these results.
Conflict of Interest
The authors declares no conflict of interest.
Funding Source
This study was funded in part by The Can Tho University Improvement Project VN14-P6, Supported by a Japanese ODA loan.
References
- Apun K., Yusof A. M., Jugang K. Distribution of bacteria in tropical freshwater fish and ponds. Int J Environ Health Res. 1999; 9(4):285-292. DOI: https://doi.org/10.1080/09603129973083.
- Pal D., Gupta C. D. Microbial pollution in water and its effect on fish. J Aquat Anim Health. 1992; 4(1):32-39. DOI: https://doi.org/10.1577/1548-8667(1992)004.
- Guzmán M. C., de los Angeles Bistoni M., Tamagnini L. M., González R. D. Recovery of Escherichia coli in fresh water fish, Jenynsia multidentata and Bryconamericus iheringi. Water Res. 2004; 38(9):2368-2374. DOI: https://doi.org/10.1016/j.watres.2004.02.016.
- Tong Thi A. N., Noseda B., Samapundo S., Nguyen B. L., Broekaert K., Rasschaert G., Heyndrickx M., et al. Microbial ecology of Vietnamese Tra fish (Pangasius hypophthalmus) fillets during processing. Int J Food Microbiol. 2013; 167(2):144-152. DOI: https://doi.org/10.1016/j.ijfoodmicro.2013.09.010.
- Little D. C., Bush S. R., Belton B., Phuong N. T., Young J. A., Murray F. J. Whitefish wars: Pangasius, politics and consumer confusion in Europe. Marine Policy. 2012; 36(3):738-745. DOI: https://doi.org/10.1016/j.marpol.2011.10.006.
- ICMSF. The International Commission on Microbiological Specifications for Foods. Micro-organisms in Food 6: Microbiological Ecology of Food Commodities. New York: Kluwer Academic/Plenum Publishers; 2005.
- Sarter S., Nguyen H. N. K., Lazard J., Montet D. Antibiotic resistance in Gram-negative bacteria isolated from farmed catfish. Food Control. 2007; 18(11):1391-1396. DOI: https://doi.org/10.1016/j.foodcont.2006.10.003.
- Woraprayote W., Pumpuang L., Tosukhowong A., Zendo T., Sonomoto K., Benjakul S., Visessanguan W. Antimicrobial biodegradable food packaging impregnated with Bacteriocin 7293 for control of pathogenic bacteria in Pangasius fish fillets. LWT – Food Science and Technology. 2018; 89(2018):427-433. DOI: https://doi.org/10.1016/j.lwt.2017.10.026.
- Hammerum A. M., Heuer O. E. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin Infect Dis. 2009; 48(7):916-921. DOI: https://doi.org/10.1086/597292.
- Hunter P. A., Dawson S., French G. L., Goossens H., Hawkey P. M., Kuijper E. J., Nathwani D., et al. Antimicrobial-resistant pathogens in animals and man: prescribing, practices and policies. J Antimicrob Chemother. 2010; 65(1):3-17. DOI: https://doi.org/10.1093/jac/dkp433.
- Jiang H. X., Tang D., Liu Y. H., Zhang X. H., Zeng Z. L., Xu L., Hawkey P. M. Prevalence and characteristics of β-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J Antimicrob Chemother. 2012; 67(10):2350-2353. DOI: https://doi.org/10.1093/jac/dks250.
- World Fishing and Aquaculture. Vietnam targets US$7bn fisheries exports. David Hayes looks at how Vietnam is planning to boost its fisheries exports (16 Jun 2016), Available at: https://www.worldfishing.net/news101/regional-focus/vietnam-targets-us$7bn-fisheries-exports. Accessed Date: 16 July 2020; 2016.
- Heuer O. E., Kruse H., Grave K., Collignon P., Karunasagar I., Angulo F. J. Human health consequences of use of antimicrobial agents in aquaculture. Clin Infect Dis. 2009; 49(8):1248-1253. DOI: https://doi.org/10.1086/605667.
- Tong Thi A. N. Assessment of antibiotic resistance and bacterial contamination of ice sold in Cantho city, Vietnam. Vietnam Journal of Science and Technology. 2019; 57(3B):49-58. DOI: https://doi.org/10.15625/2525-2518/57/3B/14205.
- CLSI. Clinical and Laboratory Standards Institute, 29th Edition, M100 – Performance Standards for Antimicrobial Susceptibility Testing. In. Clinical and Laboratory Standards Institute. National Commitee for Clinical Laboratory Standards, Wayne, PA, January 2019; 2019.
- Novoslavskij A., Terentjeva M., Eizenberga I., Valciņa O., Bartkevičs V., Bērziņš A. Major foodborne pathogens in fish and fish products: a review. Ann Microbiol. 2016; 66(1):1-15. DOI: https://doi.org/10.1007/s13213-015-1102-5.
- Nayak S. K. Role of gastrointestinal microbiota in fish. Aquac Res. 2010; 41(11):1553-1573. DOI: https://doi.org/10.1111/j.1365-2109.2010.02546.x.
- Xia J. H., Lin G., Fu G. H., Wan Z. Y., Lee M., Wang L., Liu X. J., et al. The intestinal microbiome of fish under starvation. BMC Genomics. 2014; 15(266):1-11. DOI: https://doi.org/10.1186/1471-2164-15-266.
- Mente E., Nikouli E., Antonopoulou E., Martin S. A., Kormas K. A. Core versus diet-associated and postprandial bacterial communities of the rainbow trout (Oncorhynchus mykiss) midgut and faeces. Biology Open. 2018; 7(6):34. DOI: https://doi.org/10.1242/bio.034397.
- Yagoub S. O. Isolation of Enterobacteriaceae and Pseudomonas spp. from raw fish sold in fish market in Khartoum state. Journal of Bacteriology Research. 2009; 1(7):85-88. DOI: https://doi.org/10.5897/JBR.9000032.
- Salako D., Trang P. N., Ha N. C., Miyamoto T., Ngoc T. T. A. Prevalence of antibiotics resistance Escherichia coli collected from Pangasius catfish (Pangasius hypophthalmus) fillets during processing at two factories in Mekong Delta Vietnam. Food Research. 2020; 4(5):1785-1793. DOI: https://doi.org/10.26656/fr.2017.4(5).160.
- Schjørring S., Krogfelt K. A. Assessment of bacterial antibiotic resistance transfer in the gut. International Journal of Microbiology. 2011. DOI: https://doi.org/10.1155/2011/312956.
- RASFF. Rapid alert notifications for food and feed. Available at: https://webgate.ec.europa.eu/rasff-window/portal/?event=notificationsList&StartRow. Accessed Date: 02 March 2020; 2020.
- Boss R., Overesch G., Baumgartner A. Antimicrobial resistance of Escherichia coli, Enterococci, Pseudomonas aeruginosa, and Staphylococcus aureus from raw fish and seafood imported into Switzerland. J Food Prot. 2016; 79(7):1240-1246. DOI: https://doi.org/10.4315/0362-028X.JFP-15-463.
- Nguyen D. T. A., Kanki M., Do Nguyen P., Le H. T., Ngo P. T., Tran D. N. M., Le N. H., et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int J Food Microbiol. 2016; 236:115-122. DOI: https://doi.org/10.1016/j.ijfoodmicro.2016.07.017.
- Angulo F., Nargund V., Chiller T. Evidence of an association between use of anti‐microbial agents in food animals and anti‐microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. Journal of Veterinary Medicine. 2004; 51(8‐9):374-379. DOI: https://doi.org/10.1111/j.1439-0450.2004.00789.x.
- Asai T., Kojima A., Harada K., Ishihara K., Takahashi T., Tamura Y. Correlation between the usage volume of veterinary therapeutic antimicrobials and resistance in Escherichia coli isolated from the feces of food-producing animals in Japan. Jpn J Infect Dis. 2005; 58(6):369-372.
- Levy S. B., Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nature Medicine. 2004; 10(12):122-129. DOI: https://doi.org/10.1038/nm1145.
- Campos J., Gil J., Mourão J., Peixe L., Antunes P. Ready-to-eat street-vended food as a potential vehicle of bacterial pathogens and antimicrobial resistance: an exploratory study in Porto region, Portugal. Int J Food Microbiol. 2015; 206(2015):1-6. DOI: https://doi.org/10.1016/j.ijfoodmicro.2015.04.016.
- Kahlmeter G., Menday P., Cars O. Non-hospital antimicrobial usage and resistance in community-acquired Escherichia coli urinary tract infection. J Antimicrob Chemother. 2003; 52(6):1005-1010. DOI: https://doi.org/10.1093/jac/dkg488.
- Le T. X., Munekage Y., Kato S. Antibiotic resistance in bacteria from shrimp farming in mangrove areas. Sci Total Environ. 2005; 349(1-3):95-105. DOI: https://doi.org/10.1016/j.scitotenv.2005.01.006.
- Hoa P. T. P., Managaki S., Nakada N., Takada H., Shimizu A., Anh D. H., Viet P. H., et al. Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci Total Environ. 2011; 409(15):2894-2901. DOI: https://doi.org/10.1016/j.scitotenv.2011.04.030.
- Szmolka A., Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Frontiers in Microbiology. 2013; 4(2013):258. DOI: https://doi.org/10.3389/fmicb.2013.00258.
- Maynard C., Fairbrother J. M., Bekal S., Sanschagrin F., Levesque R. C., Brousseau R., Masson L., et al. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149: K91 isolates obtained over a 23-year period from pigs. Antimicrob Agents Chemother. 2003; 47(10):3214-3221. DOI: https://doi.org/10.1128/aac.47.10.3214-3221.2003.
- Ariza R., Cohen S., Bachhawat N., Levy S., Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994; 176(1):143-148. DOI: https://doi.org/10.1128/jb.176.1.143-148.1994.

This work is licensed under a Creative Commons Attribution 4.0 International License.