Introduction
Generally, people rarely consume raw milk. However, there are some people who prefer consuming natural, unprocessed food. These people believe that raw unpasteurized milk, which has not been subject to any heating process, has particular healthy properties which reduces susceptibility to allergies, enhances nutritional quality, and has a better taste1,2.
A sound sanitary manner is needed for the production and processing of wholesome and nutritious food according to consumer’s preference. Milk is a nutrient dense white fluid secreted by female mammary gland and widely consumed food among all age groups. Quality milk convey the meaning of normal chemical composition, being completely free of harmful bacteria and harmful toxic substances, free of sediment and extraneous substances, having lower degree of titratable acidity, having good flavour, being adequate in preserving quality, and having low bacterial counts. In Bangladesh, cattle rearing, milk production and distribution are mostly done by following traditional methodModern technologies for cattle rearing, machine milking process are not common practice other than big farms3,4.
The factors associated with contamination of raw milk are the milking machine, milking area, faecal contamination, personal hygiene, poor storage condition5. Dairy farms mainly use antimicrobials because of the intra-mammary inflammation2. Uncontrolled usage of antimicrobial components may affect negatively on human health by residing the residues in cattle body. Whereas controlled usage of antimicrobials can help to rear healthy cattle which would be added value in dairy industry6.
Previously, Tekilegiorgis mentioned about a study in Ethiopia in 2018 where total bacterial count was 5×103 to 3.18×108 cfu/ml in raw milk and 4.4×101 to 4.43×105 cfu/ml in pasteurized milk samples7. Regasa et al., (2019) reported about Staphylococcus aureus susceptibility was 16.6% and load count was 104-105 cfu/ml 8 In Northern Italy (2016), the prevalence of Listeria monocytogenes was reported as 1.66%, more specifically 2.2% from bulk milk tank whereas 0.5% in vending machine of milk9. Ahmed et al., (2019) reported about five different pasteurized brands of milk from Bangladesh which ranged from 3.5×104 to 1.15×107 cfu/ml. According to Bangladesh Standards and Testing Institution, the total bacterial count should not exceed 20,000 CFU/ml10. In 2015, raw, pasteurized milk and yogurt samples were collected from different zones of Dhaka city, Bangladesh. In that report, it has been mentioned that total viable count varied from 3.5×103 cfu/ml to 4.2×106 cfu/ml for raw milk samples. Along with this, all of them were contaminated with E. coli and Shigella-like species but Listeria was not present11. Previously, standard bacterial plate count for pasteurized milk was reported in Sylhet city, Bangladesh where the range was also higher than the recommended range (54200 to 68400 cfu/ml)12.
In addition to this issue, some organisms are present as potential for food borne illnesses and some of them are comprised of genes which are antimicrobial resistant. They are also mentioned for the mycotoxin and presence of their metabolites in the milk and dairy products13. Higher level of yeast and lower level of mould compared to yeast had been reported in different studies on raw milk14,15.
As milk is an ideal media for growth of microorganisms, it is very crucial to investigate the microbial contamination load and associated microorganisms’ presence in milk16. Although most of the cases milk is pasteurized before marketing, microorganisms can be a vital concern regarding health aspects, as there are several situations when pasteurized milk cannot be helpful15.
In several researches it has been mentioned that concern in the dairy industry has raised because of the disease outbreaks from the consumption of unpasteurized milk by farm employees, family members, associated neighbours and nearby local area population. Unpasteurized milk is also used in the cheese industry. Along with this, contamination in the milk processing industry also allow forming of biofilms and some improper pasteurization may not abolish food borne microorganisms2. No previous data was found on the microbial quality assessment of milk in this area. As milk is widely consumed food among all age groups thus the present study aims to identify the presence of pathogenic microorganisms in raw milk and antibiotic resistance of those organisms in the mentioned region.
Materials and Methods
Sample Preparation
Five raw milk samples were collected from five different locations of Noakhali, Bangladesh. Two different brands of UHT milk samples were collected from the same region. Selection of collection zones and brands was done randomly. The samples were labelled as S1, S2, S3, S4, S5 and UHT and those were collected from University student hall, Suborno-agro, Sonapur, Maijdee, VC- Bungalow and local shops, respectively. All of them were collected and transported in ice box to the laboratory of Food Technology and Nutrition Science, Department of Noakhali Science and Technology University. Then samples were stored in the laboratory at 4 ºC. 1 ml of milk sample was mixed with 9ml of 0.9% sterile sodium chloride solution in a sterilized cotton plugged test tube. Then it was mixed by stirring and shaking and this homogenized solution was then allowed for further serial dilution. The method was followed from Mokbul et al. (2016)17.
Bacteriological Studies
For the isolation of bacteria, pour plate and streak plate techniques were followed. Nutrient Agar (NA) and MacConkey (MCA), Eiosin Methylene Blue (EMB) and Genital menital salt agar (GMSA) were used for isolation purpose. Nutrient agar was used for cultivating non-fastidious microorganisms. MacConkey agar was used for the isolation and differentiation of enteric bacteria. EMB and GMSA agar are highly selective media and they were used for isolating E. coli and Staphylococcus. All these media were prepared according to their manual. 10 folds dilution was done to reduce the density of the microorganisms. Pour plate technique was used and then it was incubated for 24 hours at 370C that were grown in NA, MCA, EMB and GMSA. Colonies were isolated and collected based on their color, shape, elevation and stored in the nutrient agar slant. Morphological and cultural tests were done immediately.
Isolate Identification
Biochemical characterization as Kliger’s Iron Agar (KIA) test, Motility Indole Urease (MIU) test, Catalase and Oxidase tests were performed for bacterial identification. The procedures were followed from Mokbul et al., (2016)17.
Antibiogram Profiling
Isolated strains were inoculated, prepared in Mueller-Hington broth and adjusted to turbidity equal to 0.5 McFarland standards and were applied onto Mueller-Hinton agar using a wire loop. Sterilized swab was then used to spread the culture on the media. The inoculated plate was allowed to dry for a few minutes, after which sensitivity disks were applied to it using sterile forceps. Zones of inhibition around sensitivity disks were measured after 18-24hr of incubation at 37°C. The sensitivity of all isolates was tested against: Rifampcin (RIF) 5μg/disk, Cefotaxime(CTX)30μg/disk, Amikacin(AK)30μg/disk, Colistin(CL)10μg/disk, Genetamicin (Gen)10μg/disk, Chloramphenicol(c)30μg/disk, Ciprofloxacin(CIP)15μg/disk, Aoxycillin(AMX)30μg/disk, Ceflriaxone(CTR)30μg/disk, Kanamycin(K)30μg/disk, Nitrofurantoin(NIT)30μg/disk, Norfloxacin(NX)10μg/disk, and Cefepime (CPM) 30 μg/disk according to the CLSI requirements using the disk diffusion method.
The interpretation of zones of inhibition around the disks was done according to CLSI (2006) (American Public Health Association, 1913).
Statistical Analysis
SPSS software version 23.0 was used to perform the Analysis of Variance (ANOVA) test in order to understand the significant difference between different samples. The level of significance was set at ≤0.05.
Result and Discussion
The study revealed that all raw milk samples were contaminated and in certain cases, pathogens were detected which is a public health concern. The bacterial load of all the samples was quite high (Table 1) and in commercial UHT milk, no microorganisms were found. The range of the bacterial load of raw milk samples was found in different agar as Nutrient agar 1.95×104 to 1.87×106, Macconkey agar 7.5×105 to 1.95×106, Eosin methylene blue agar 2.33×104 to 1.35×106, and Glucose minimal salt agar 1.0×102 to 1.70×106. Among all the samples, the sample from Maijdee Bazar (S4) contained the highest bacterial load whereas the sample from VC-bungalow (S5) contained the lowest bacterial count. According to the Bureau of Indian Standards (BIS) for raw milk plate count (SPC) (IS: 1479-1977, PART 111) if the bacterial count is (count/ml) greater than 200,000 then the milk is of better quality, if the count is from 2,000,01 to 1,000,000 then it is of good quality, 1,000,000 to 50, 00,000 is Fair, and if it is more than 5,000,000 then it is called poor quality milk18. Hence comparing this study with standards, S5 quality was best among all, S3 was better quality and S1 and S4 showed the fair quality of milk-based on nutrient agar total viable count. According to statistical results, colony count varied significantly in NA for S1 compared to S3, S4 and S5. In MCA, S1, S2 and S3 varied significantly among each other.
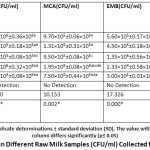 |
Table 1: Bacterial Count in Different Raw Milk Samples (CFU/ml) Collected from Different Locations. |
Biochemical characterization tests were performed to identify the microorganisms in raw milk samples. After conducting the tests, E. coli, Salmonella, Staphylococcus, Bacillus, Listeria monocytogens, and Yersenia were identified in the samples (Table 2). About half (47%) of the identified microorganisms were E. coli and only 5.88% were Bacillus, Listeria and Yersenia (Table 3). Previously, microbial contamination assessment had been reported for raw, pasteurized and UHT milk. Amenu et al. (2019) reported 2.5% E. coli contamination in milk and milk products samples in Ethiopia19. E. coli contamination had been reported in Italy in 2009 in vending machine as well. They revealed 0.2% E. coli, 0.3% Salmonella spp., 1.5% Campylobacter spp., and 1.6% Listeria monocytogens contamination in all the samples20. In 2011 Hossain et. al., analyzed samples of raw, pasteurized and UHT milk from twelve different local markets of different locations in Bangladesh. They concluded that most of the raw milk samples contained indicator and pathogenic organisms as coliform, Aeromonas, Salmonella, and Staphylococcus. Some raw and pasteurized milk also contained psychrophilic organisms4. In 2019, one study reported 10.8% S. aureus harbor in ready-to-consume raw milk and milk products in Ethopia19. Huque et al., (2018) mentioned about the total bacterial count in raw milk as 2.31 x 105 to 2.45 x 105 CFU/ml in Savar, Bangladesh21. In different zones of Dhaka, Bangladesh, total bacterial count varied between 4.2×106 to 3.5 × 103 CFU/ml11. In another review by Zastempowska et. al., (2016) it was mentioned that many societies consume raw milk and Salmonella, Shiga toxin-producing E. coli. Micobacterium bravis, Campylobacter were responsible for the disease outbreaks in many cases13. Bianchi et al., (2009) mentioned in a similar statement that unpasteurized milk can be a possible source of food-borne disease outbreak for many organisms20. All the organisms that are reported in this study are in agreement with the previous reports from various researchers of the world.
Table 2: Biochemical Tests and Identification of Microorganisms in Raw Milk Samples.
| Isolates ID | KIA Test | MIU Test | H2s production | Citrate test | Identification | ||||
| Slant | Lactose | Gas | Motility | Indole | Urease | ||||
| M/E/D | K | – | + | – | – | – | – | – | Yersenia |
| M/E/R | K | – | – | – | – | – | + | – | Salmonella |
| M/E/G | A | + | + | + | + | – | – | – | E. coli |
| M/M/P | A | + | + | + | + | – | + | – | E. coli |
| M/M/R | A | + | + | + | + | – | + | – | E. coli |
| M/E/P | A | + | + | + | + | – | + | – | E.coli |
| S/E/G | A | + | + | + | + | – | + | – | E.coli |
| V/E/P | K | – | – | – | – | – | + | – | Salmonella |
| V/M/R | A | + | + | + | + | – | + | – | E.coli |
| Su/E/G | A | + | + | + | + | – | + | – | E.coli |
| Su/E/P | K | – | – | – | – | – | + | – | Salmonella |
| H/E/P | K | – | – | – | – | – | + | – | Salmonella |
| H/E/G | A | + | + | + | + | – | + | – | E.coli |
| H/N/P | A | + | – | + | – | – | – | + | Listeria monocytogens |
| V/N/P | A | + | – | + | – | – | – | + | Bacillus |
| M/N/P | A | + | – | – | – | + | – | + | Staphylococcus |
| S/N/P | A | + | – | – | – | + | – | + | Staphylococcus |
Isolate ID: Sample/Media/Colour; (+) means positive; (-) means negative
Table 3: Percentage of Isolates in the Samples.
| Name of the microorganism | N (%) |
| E.coli | 8(47.05) |
| Salmonella | 4(23.53) |
| Staphylococcus | 2(11.76) |
| Bacillus | 1(5.88) |
| Listeria monocytogens | 1(5.88) |
| Yersenia | 1(5.88) |
After performing the biochemical test, E. coli, Salmonella, Staphylococcus, Bacillus, Listeria monocytogens and Yersinia were detected. Similarly, Rahman et al., (2015) mentioned E. coli, Salmonella, Listeria in raw milk samples in Dhaka city, Bangladesh11.
Figure 1 shows the antibiotic resistance study of the isolates. This examination has been done with six different isolates and they showed different resistance for different antibiotics.
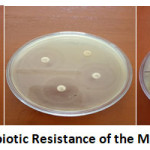 |
Figure 1: Antibiotic Resistance of the Microorganisms. |
The antimicrobial resistance profiles of the bacterial isolates from raw cow milk are summarized in Table 4. All of the isolates showed antibiotic resistance though the MRI% varied organism to organism.
In this study, Staphylococcus was sensitive to RIF, CTX, AK, CL, Gen, C, CIP, AMX, K, NIT and NX. A similar finding was reported by Pol and Ruegg (2007) and Frey et al., (2013). They mentioned coagulase-negative Staphylococci from bovine milk were resistant to oxacillin, streptomycin, erythromycin, kanamycin, gentamycin22,23. E. coli showed intermediate resistance and completely resistant to CTR, CPM, K, RIF, CTX, AMX. Among these, Cefoxitin, Ceftriaxone, Kanamycin resistance were in agreement with previously reported research by24. The MRI percentage for E. coli, Salmonella and Listeria were 23%, 38.46% and 46.15%, respectively. For these three organisms, multi-drug resistance was reported by Obaidat and Stringer (2019) in Jordan. They mentioned a higher percentage of resistance as 93.8, 79.2, and 57.1 for L. monocytogenes, E. coli and S. enterica, respectively. Both Listeria and E. coli were resistant to RIF CTX and AMX whereas Salmonella was resistant to RUF, CTX, CIP, NX and CPM. Kanamycin was in an intermediate resistance level for both E. coli and Salmonella, which was also mentioned by Obaidat and Stringer (2019) in the resistant category25. Yersinia was intermediate level resistant to CIP, CTR and CPM and resistant to RIF, CTX and AMX. These findings were similar to the study conducted by Bonardi et al., (2018). They also mentioned ciprofloxacin, nalidixic acid, ceftriaxone, tetracycline, ticarcillin as being sensitive to, and amoxicillin, cefoxitin, cephalexin as in the resistant category for Yersinia26.
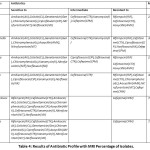 |
Table 4: Results of Antibiotic Profile with MRI Percentage of Isolates. |
Conclusion
Considering bacterial load in raw milk, it was observed that VC-bungalow (S5) contained the lowest bacterial count and Maijdee Bazar (S4) contained the highest bacterial load which quality was labeled as fair compared with the standard bacterial load. No bacterial presence was recorded in UHT milk. Along with E. coli other microorganisms as Salmonella, Listeria, Bacillus, Yersinia and Staphylococcus presence were observed. Other than Staphylococcus and Bacillus, all of them were resistant to three or more antibiotics which are alarming in the global health aspects as well. However, at this age of globalization and commercialization, antibiotic resistance will affect country borders. So, this should be taken care of on a high-priority basis. It is worth mentioning that integrated monitoring and surveillance of the usage of different antibiotics for cattle is required. Proper education among farmers and throughout the community about the after-effect of antibiotic resistance is important as well to regulate the situation.
Acknowledgment
The present study was conducted in the laboratory of the Department of Food Technology and Nutrition Science, Noakhali Science and Technology University, Bangladesh. The authors wish to thank the chairman of the department for work permission and the technical assistants for their support throughout the work.
Funding Sources
The authors received no financial support for this research, authorship or publication.
Conflict of Interest
The authors have no conflict of interest.
References
- Claeys WL, Cardoen S, Daube G, et al. Raw or heated cow milk consumption: Review of risks and benefits. Food control 2013;31(1):251-62.
CrossRef - Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathogens & Disease 2005;2(2):115-29.
CrossRef - Ghosh S, Sharmin F, Mokbul M, Kabir MR, Alam MR. Physiochemical Quality of Locally Packaged Raw Milk of Noakhali Region, Bangladesh Noakhali Science and Technology University, 2020.
- Hossain T, Alam M, Sikdar D. Chemical and microbiological quality assessment of raw and processed liquid market milks of Bangladesh. 2011.
CrossRef - Boor KJ, Wiedmann M, Murphy S, Alcaine S. A 100-year review: microbiology and safety of milk handling. Journal of dairy science 2017;100(12):9933-51.
CrossRef - Erginkaya Z, Turhan E, Tatlı D. Determination of antibiotic resistance of lactic acid bacteria isolated from traditional Turkish fermented dairy products. Iranian journal of veterinary research 2018;19(1):53.
- Tamirat T. Microbiological quality analysis of raw and pasteurized milk samples collected from Addis Ababa and its surrounding in Ethiopia. Approaches in Poultry, Dairy and Veterinary Sciences 2018;4(5):374-81.
- Regasa S, Mengistu S, Abraha A. Milk Safety Assessment, Isolation, and Antimicrobial Susceptibility Profile of Staphylococcus aureus in Selected Dairy Farms of Mukaturi and Sululta Town, Oromia Region, Ethiopia. Veterinary medicine international 2019;2019: 1-11.
CrossRef - Dalzini E, Bernini V, Bertasi B, Daminelli P, Losio M-N, Varisco G. Survey of prevalence and seasonal variability of Listeria monocytogenes in raw cow milk from Northern Italy. Food Control 2016;60:466-70.
CrossRef - Ahmed S, Zim A, Rahman S, Ghosh S, Chhetri A, Ali M. Quality and Safety Assessment of Bangladeshi Pasteurized Milk. Journal of food quality and hazards control 2019.
CrossRef - Rahman T, Akon T, Sheuli IN, Hoque N. Microbiological analysis of raw milk, pasteurized milk and yogurt samples collected from different areas of Dhaka city, Bangladesh. Journal of Bangladesh Academy of Sciences 2015;39(1):31-36.
CrossRef - Saha S, Ara A. Chemical and Microbiological Evaluation of Pasteurized Milk Available in Sylhet City of Bangladesh. The Agriculturists 2012;10(2):104-08.
CrossRef - Zastempowska E, Grajewski J, Twarużek M. Food-borne pathogens and contaminants in raw milk–a review. Annals of Animal Science 2016;16(3):623-39.
CrossRef - Lagneau P, Lebtahi K, Swinne D. Isolation of yeasts from bovine milk in Belgium. Mycopathologia 1996;135(2):99-102.
CrossRef - Quigley L, O’Sullivan O, Stanton C, et al. The complex microbiota of raw milk. FEMS microbiology reviews 2013;37(5):664-98.
CrossRef - Gopal N, Hill C, Ross PR, Beresford TP, Fenelon MA, Cotter PD. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Frontiers in microbiology 2015;6:1418.
CrossRef - Mokbul M, Islam T, Alim SR. Bacteriological Quality Analysis of Ice Cream Produced by the Small Factories of Dhaka City. International Journal of Health Sciences and Research 2016;6:235-40.
- Pant R, Nirwal S, Rai N. Prevalence of antibiotic-resistant bacteria and analysis of microbial quality of raw milk samples collected from different regions of Dehradun. International Journal of PharmTech Research 2013;5(2):804-10.
- Amenu K, Grace D, Nemo S, Wieland B. bacteriological quality and safety of ready-to-consume milk and naturally fermented milk in Borana Pastoral Area, Southern Ethiopia. Tropical animal health and production 2019;51(7):2079-84.
CrossRef - Bianchi DM, Barbaro A, Gallina S, et al. Monitoring of foodborne pathogenic bacteria in vending machine raw milk in Piedmont, Italy. Food Control 2013;32(2):435-39.
CrossRef - Huque R, Hossain A, Jolly Y, et al. Evaluation of elemental, microbial and biochemical status of raw and pasteurized cow’s milk. International Food Research Journal 2018;25(4):1682-90.
- Pol M, Ruegg P. Relationship between antimicrobial drug usage and antimicrobial susceptibility of gram-positive mastitis pathogens. Journal of dairy science 2007;90(1):262-73.
CrossRef - Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. Journal of dairy science 2013;96(4):2247-57.
CrossRef - Saini V, McClure J, Scholl DT, DeVries TJ, Barkema HW. Herd-level relationship between antimicrobial use and presence or absence of antimicrobial resistance in gram-negative bovine mastitis pathogens on Canadian dairy farms. Journal of dairy science 2013;96(8):4965-76.
CrossRef - Obaidat MM, Stringer AP. Prevalence, molecular characterization, and antimicrobial resistance profiles of Listeria monocytogenes, Salmonella enterica, and Escherichia coli O157: H7 on dairy cattle farms in Jordan. Journal of dairy science 2019;102(10):8710-20.
CrossRef - Bonardi S, Le Guern A, Savin C, et al. Detection, virulence and antimicrobial resistance of Yersinia enterocolitica in bulk tank milk in Italy. International dairy journal 2018;84:46-53.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.







